Qc tab – Bio-Rad Firmware & Software Updates User Manual
Page 131
Users and Preferences
120
Specify the default settings for a new Gene Expression data file:
• Relative to. Select a control or zero. To graph the gene expression data originating at 1
(relative to a control), select Control. When you assign a control sample in the
Experiment Setup window, the software automatically defaults to calculate the data
relative to that control. Select Relative to zero to instruct the software to ignore the
control, which is the default selection when no control sample is assigned in the
Experiment Settings window
• X-Axis. Graph the Target or the Sample on the x-axis
• Y-Axis. Graph Linear, Log 2, or Log 10 scale on the y-axis
• Scaling. Select a scaling option for the graph. Leave the graph unscaled. Alternatively,
choose a scaling option to scale to the highest value or to the lowest value
• Method. Set the default analysis mode, including normalized expression (C
q
) or
relative expression (C
q
)
• Error Bar. Select Std Dev. for standard deviation, or Std. Error Mean for the standard
error of the mean
• Error Bar Multiplier. Select the standard deviation multiplier to graph the error bars. The
default is 1. Change the multiplier to either 2 or 3
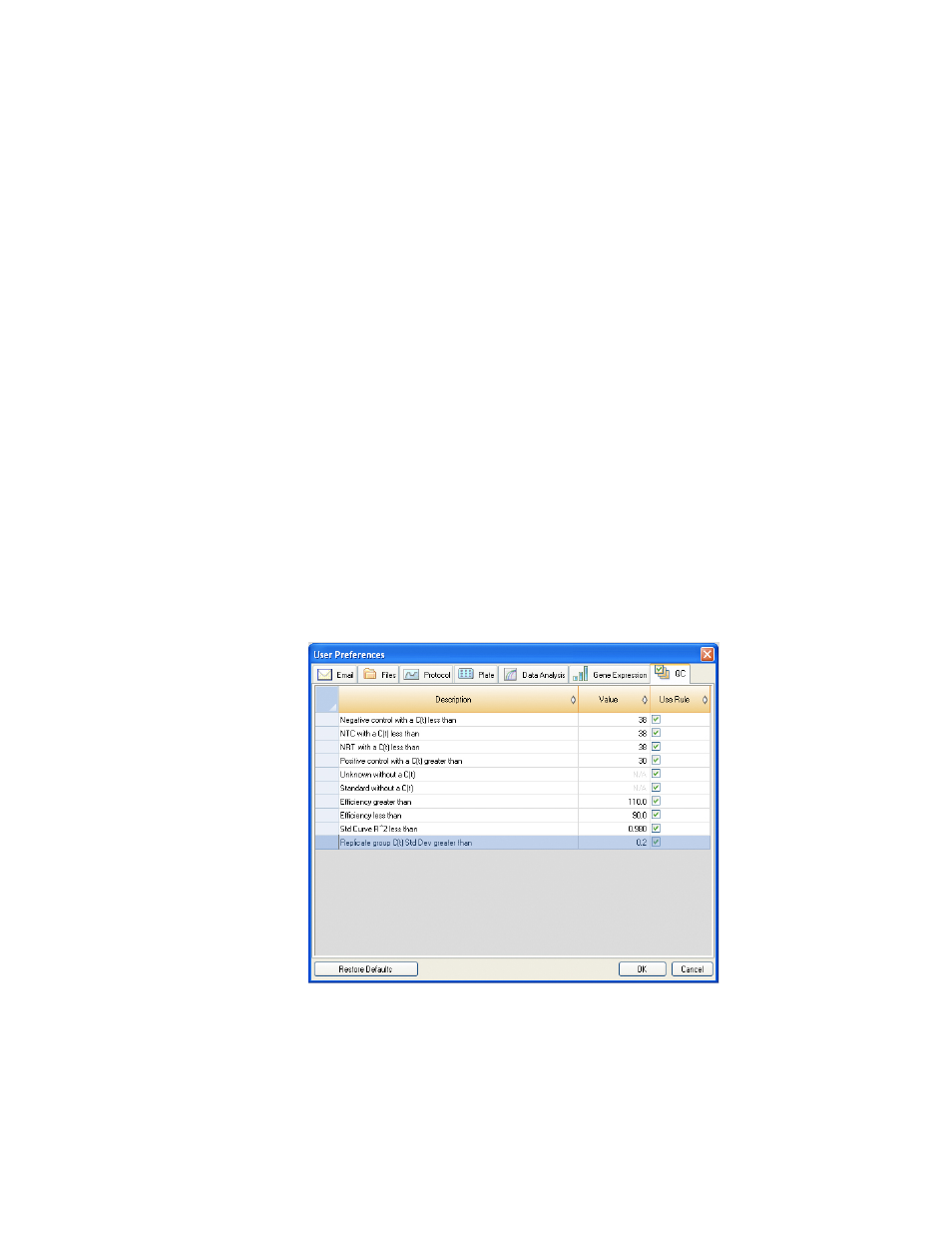
QC Tab
Select the QC tab in the User Preferences window to specify QC rules to apply to data in Data
Analysis module. The software validates the data against the enabled tests and the assigned
values.
NOTE: Wells that fail a QC parameter can easily be excluded from analysis in the
QC module of the Data Analysis Window using the right-click menu option.
Figure 81. QC tab in the User Preferences window.
Specify to add cut off values and to enable the following QC rules:
• Negative control with a C
q
less than XX. Input a C
q
Cut-off Value
• NTC (no template control) with a C
q
less than XX. Input a C
q
Cut-off Value
