Gene expression tab – Bio-Rad Firmware & Software Updates User Manual
Page 130
MiniOpticon Instruction Manual
119
For quantification data, select the following settings:
• Baseline Setting. Select the default base lining method for Analysis mode. Choose
Baseline Subtracted Curve Fit, No Baseline Subtraction, or Baseline Subtracted
• Cq Determination Mode. Select between Regression mode or Single Threshold mode
to determine how C
q
values are calculated for each fluorescence trace
• Log View. Select On to show a semi-logarithmic graph of the amplification data. Select
Off to show a linear graph
For the allelic discrimination data, select the following settings:
• Display Mode. Select RFU to show the data as a graph of the RFU, or select C
q
to show
a graph of quantification cycles
• Normalize Data. This selection is available only when RFU is selected. Select No to
show unnormalized data. Select Yes to normalize the data to the control sample
For end point data, select the following settings. Select the number of end cycles to average
when calculating the end point calculations:
• PCR. Enter a number of cycles for PCR to average the end cycles for quantification data
(default is 5)
• End Point Only Run. Enter a number of cycles for End Point Only Run to average the
end cycles for end point data (default is 2)
For melt curve data, select to detect either positive or negative peaks.
For well selector panes, select to label wells with sample type, target name, or sample name.
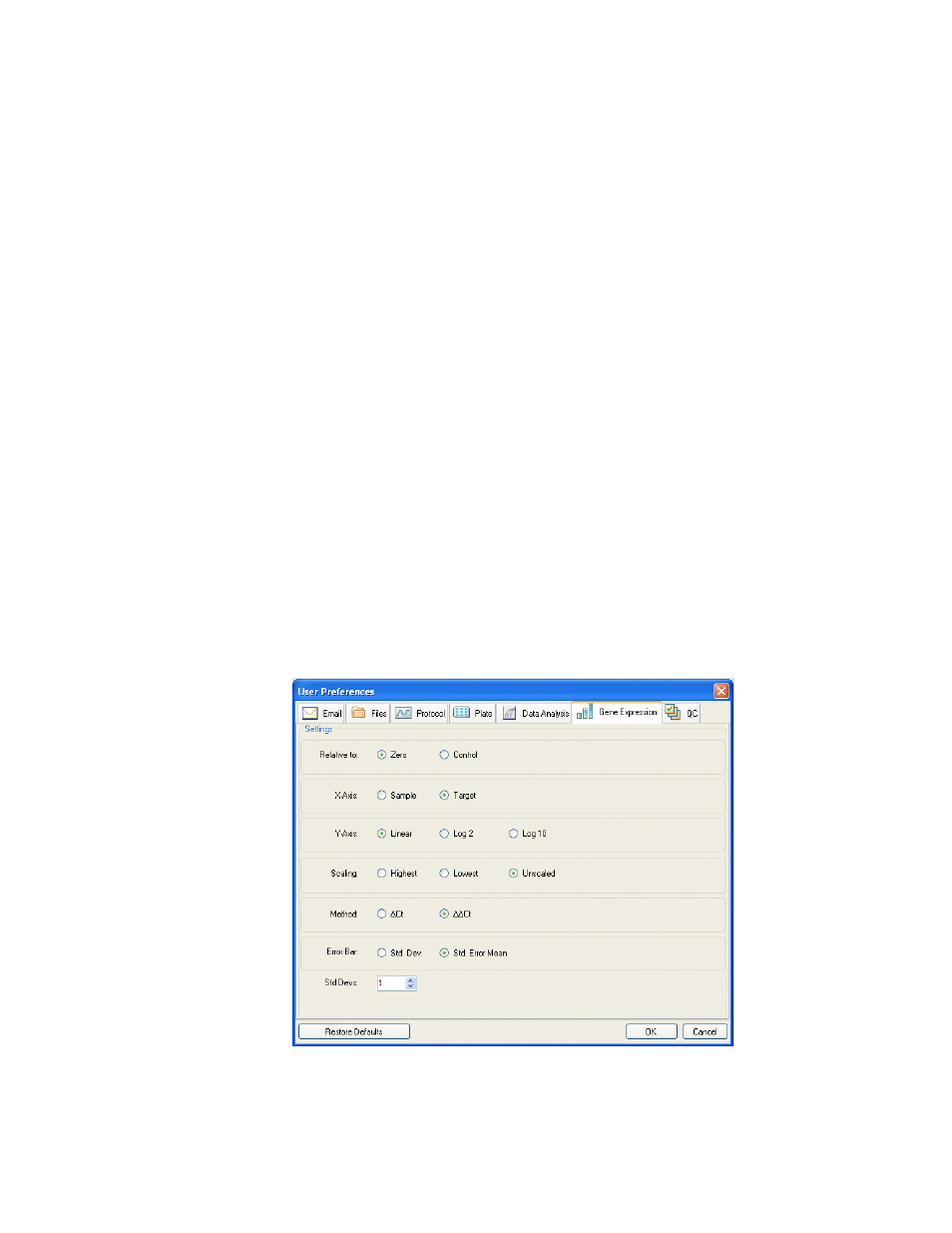
Gene Expression Tab
Select the Gene Expression tab in the User Preferences window to specify the default
settings for a new Gene Expression data file.
Figure 80. Gene Expression tab in the User Preferences window.
