2 introduction to real-time pcr, 1 pcr, 2 qualitative vs. real-time pcr – Techne PrimeQ User Manual
Page 42
42
2.2 Introduction to Real-time PCR
2.2.1 PCR
PCR is a powerful biochemical technique that has revolutionised biological research by allowing
minute amounts of DNA to be amplified millions of times in just a few hours. PCR allows the
selective amplification of a ‘target’ region of DNA lying between two specific DNA sequences
(primers). The DNA sequence lying between these primers does not need to be known, therefore
PCR allows researchers to amplify target DNA with relative ease and reproducibility.
The technique exploits the 5’ to 3’ polymerase activity of the enzyme Taq DNA polymerase
isolated from the thermophilic bacterium Thermus aquaticus. Once the primer binds to the
complementary region of the single-stranded target, the enzyme will catalyse the extension of
DNA to produce a complimentary second strand.
The primers anneal to
complementary regions on
the template DNA.
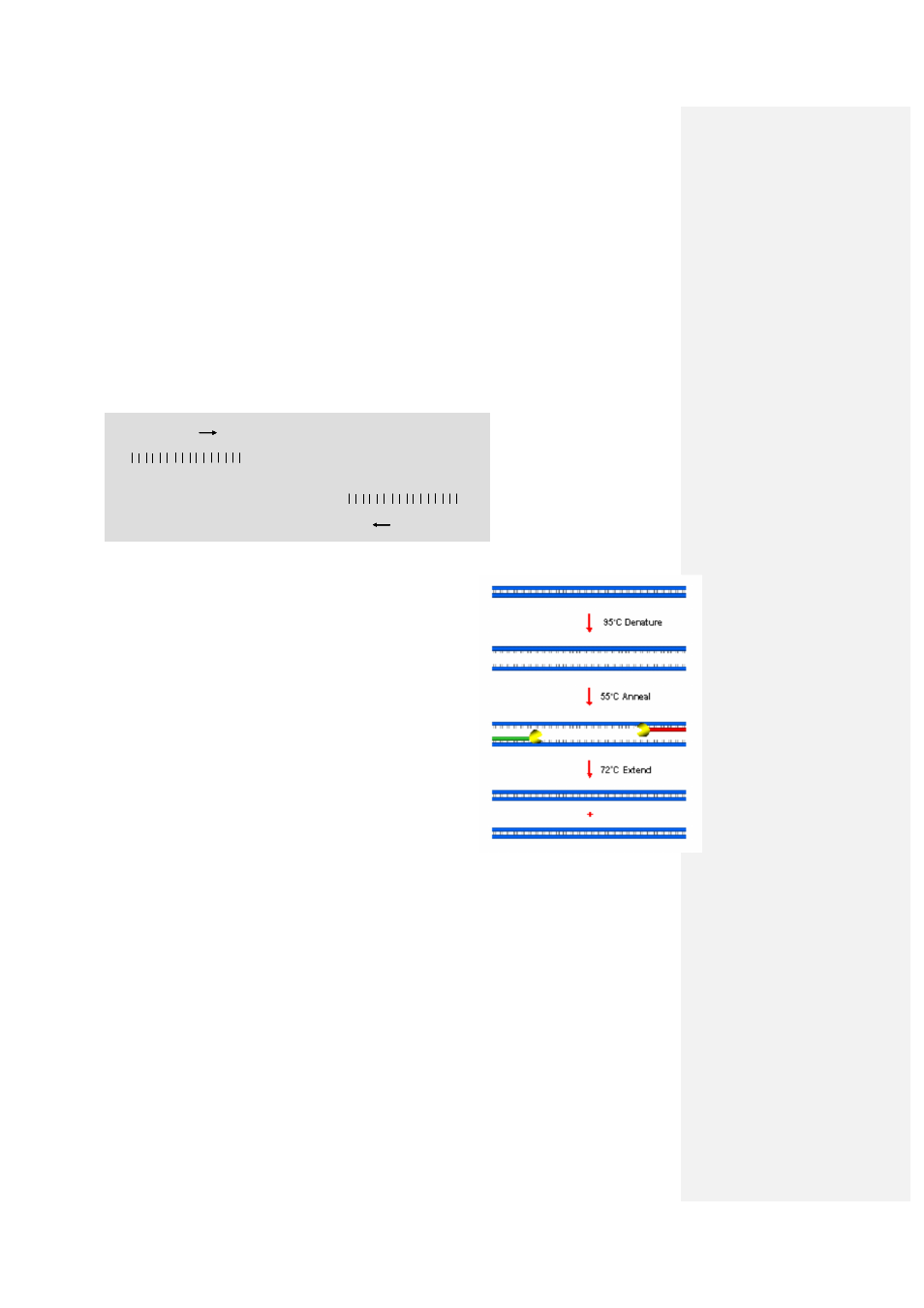
The classical PCR protocol consists of three temperature steps:
1.
Denaturation (at 95°C): In its normal state, DNA
consists of two strands made up of complementary
bases. These strands need to be separated before
the PCR can progress. The first temperature step
is therefore designed to dissociate, or denature,
these two strands.
2.
Annealing (typically between 55°C and 65°C):
This temperature step allows annealing of the
primers to complementary sequences on the
template DNA. The temperature will vary
according to the primer characteristics such as GC
content, length and sequence.
3.
Extension (72°C): When the primers have
annealed to the complementary single-stranded
DNA, the enzyme Taq DNA polymerase extends
the DNA using its 5’ to 3’ polymerase activity. The
optimal temperature for this enzyme is 72°C.
This results in the production of two new copies of the target DNA which, assuming optimal
conditions, can be amplified exponentially by repeating steps 1 to 3.
2.2.2 Qualitative vs. real-time PCR
PCR quickly became an indispensable tool for scientists wanting to amplify and characterize
genetic material. However it has one major limitation in that the results are qualitative i.e. it can
determine if a target is present but not the amount. The traditional approach to quantification was
to compare known sample concentrations of starting DNA with unknown samples cycled at a
range of concentrations and cycle numbers. The problems associated with this ‘semi-quantitative’
approach are many, including the expense of multiple PCR runs, the increased risk of
contamination through the need for downstream processing of samples and the fact that end-point
measurements have a tendency to vary between replicates. As such, the very accuracy of the
post-run method of measurement is put into question. However, real-time PCR or quantitative
A
G
C
TT
A
GG
CC
T
AA
CC
G
T
C
G
AA
T
CC
GG
A
TT
GG
C
A
G
C
T
AA
GG
TTT
CC
A
G
A
TT
CC
AAA
T
G
C
G
C
T
A
A
G
C
TT
A
GG
CC
T
AA
CC
G
T
C
G
A
TT
CC
AAA
GG
T
C
T
AA
GG
TTT
A
C
G
C
G
A
T
G
A
TT
CC
AAA
T
G
C
G
C
T
A
5’
3’
5’
3’
Forward primer
Reverse primer
A
G
C
TT
A
GG
CC
T
AA
CC
G
T
C
G
AA
T
CC
GG
A
TT
GG
C
A
G
C
T
AA
GG
TTT
CC
A
G
A
TT
CC
AAA
T
G
C
G
C
T
A
A
G
C
TT
A
GG
CC
T
AA
CC
G
T
C
G
A
TT
CC
AAA
GG
T
C
T
AA
GG
TTT
A
C
G
C
G
A
T
G
A
TT
CC
AAA
T
G
C
G
C
T
A
5’
3’
5’
3’
Forward primer
Reverse primer
A
G
C
TT
A
GG
CC
T
AA
CC
G
T
C
G
AA
T
CC
GG
A
TT
GG
C
A
G
C
T
AA
GG
TTT
CC
A
G
A
TT
CC
AAA
T
G
C
G
C
T
A
A
G
C
TT
A
GG
CC
T
AA
CC
G
T
C
G
A
TT
CC
AAA
GG
T
C
T
AA
GG
TTT
A
C
G
C
G
A
T
G
A
TT
CC
AAA
T
G
C
G
C
T
A
A
G
C
TT
A
GG
CC
T
AA
CC
G
T
C
G
AA
T
CC
GG
A
TT
GG
C
A
G
C
T
AA
GG
TTT
CC
A
G
A
TT
CC
AAA
T
G
C
G
C
T
A
A
G
C
TT
A
GG
CC
T
AA
CC
G
T
C
G
A
TT
CC
AAA
GG
T
C
T
AA
GG
TTT
A
C
G
C
G
A
T
G
A
TT
CC
AAA
T
G
C
G
C
T
A
5’
3’
5’
3’
Forward primer
Reverse primer
5’
3’
5’
3’
Forward primer
Reverse primer
