Techne PrimeQ User Manual
Page 178
178
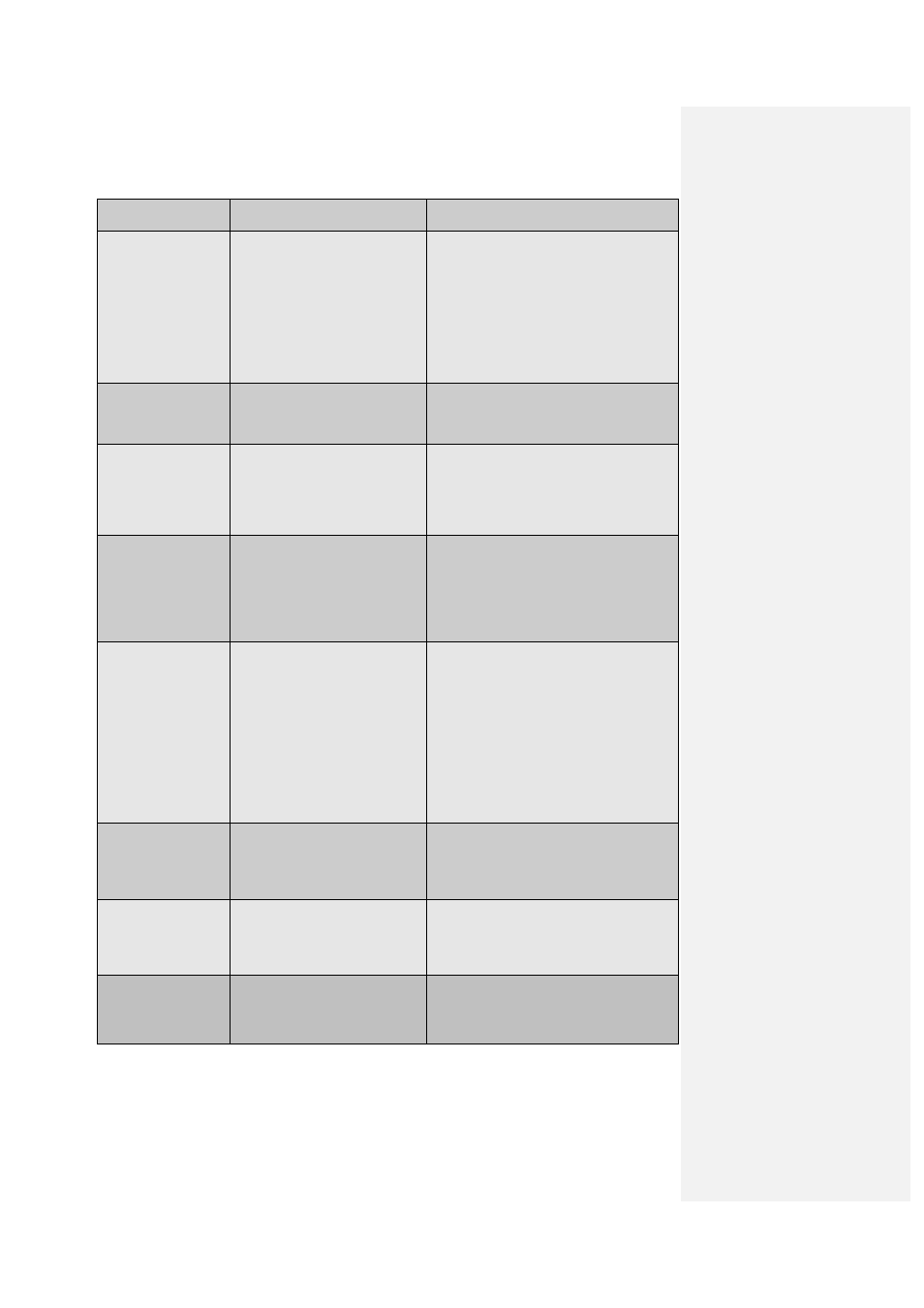
ISSUE
CAUSE
SOLUTION
No Cq recorded for
a sample.
No template added.
Repeat assay.
No target in sample.
Noise and crossing line set
incorrectly.
The default settings in Quansoft are 4
and 10 standard deviations respectively,
adjust these through the parameter
option or the Analysis Wizard.
Decreased volume
in samples at end of
a run.
Poor seal.
Ensure the sealing method is
appropriate. Heat sealing is
recommended.
Higher Cq than
expected.
Fewer templates added to
reaction.
Increase amount of template added to
reaction.
Template is degraded.
Check sample integrity on an agarose
gel or suitable bio analyser.
Lower Cq than
expected.
More template added to the
reaction.
Decrease amount of template added to
reaction.
Template or amplicon
contamination of one of the
reagents.
Repeat assay with fresh reagents.
Small increase in
fluorescence.
Concentration of dye is too low
either because the dye has
deteriorated or insufficient was
added to the reaction.
Protect reagents from light. Do not
freeze-thaw repeatedly. Check
optimization of dye concentration and
add more if necessary.
Poor PCR efficiency.
Optimize primer and probe
concentrations. Check annealing
temperatures of primers and probes.
Optimize MgCl
2
concentrations (usually
unnecessary if primers and probe were
designed correctly).
More than one peak
appears in the
dissociation curve.
Two products of different
length/GC content have been
amplified; possible causes are
miss-priming or pseudo-genes.
Check products on an agarose gel.
Optimize annealing temperature of
primers.
No specific peak
appears in the
dissociation curve,
only primer-dimers.
Assay conditions preferentially
amplify primer-dimers rather
than the specific product.
Use a hot start enzyme. Increase
annealing temperature of the primers to
increase stringency.
Amplification plots
have a flat slope.
Poor PCR efficiency.
Optimize assay conditions.
