Method guidelines, Ii. sample preparation – Waters Oligonucleotide Separation Technology XBridge OST C18 Columns User Manual
Page 3
3
[ method guidelines ]
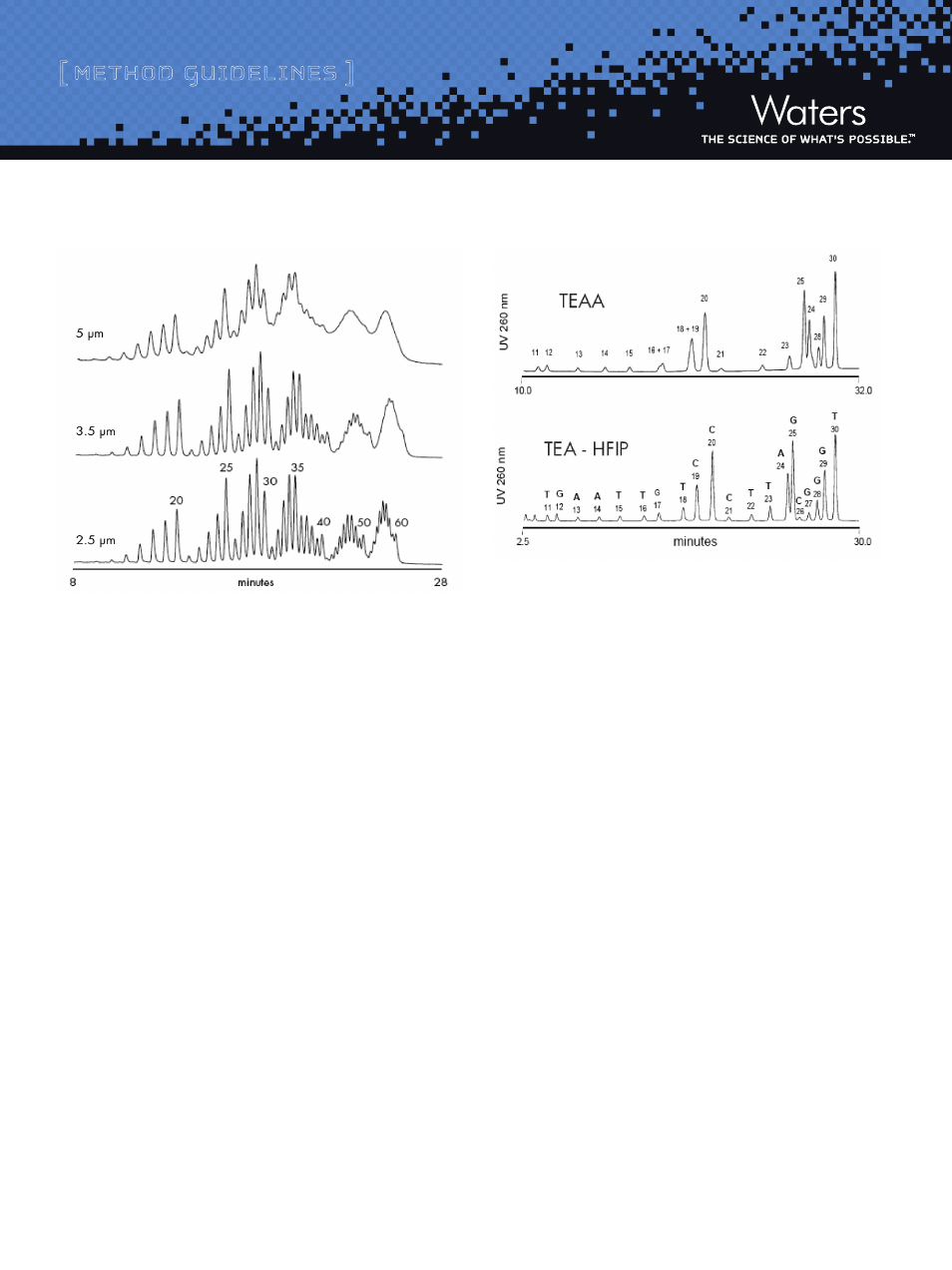
Figure 3: Effectiveness of Waters BEH Technology
™
Hybrid-Silica C
18
Particle Size on Deoxythymidine Ladder Separations
HPLC system:
Waters BioAlliance
™
2796, PDA Detector with micro UV cell
Sample Injected:
Approximately 100 pmoles of detritylated 15 – 60mer crude
oligonucleotide ladder diluted in 0.1 M TEAA
Column:
Waters BEH Hybrid-Silica C
18
particles (2.1 x 50 mm)
Mobile Phases:
A: 0.1 M TEAA,
B: Acetonitrile / 0.1M TEAA, 20/80, v/v
Flow rate:
0.2 mL/min
Column Temp.:
60 ˚C
Gradient delay:
0.45 mL
Gradient:
40 to 62.5% B in 30 minutes (8-12.5% acetonitrile,
0.15% acetonitrile per minute)
Detection:
260 nm, 5 scans per second
In addition to ion-pairing, a hydrophobic reversed-phase mechanism
also takes place in the oligonucleotide separation. The residual interaction
of nucleobases has an impact on overall retention and separation
selectivity, especially when using Triethylammonium Acetate (TEAA)
ion-pairing mobile phases. Separation of N and N-1mers may be either
enhanced or suppressed by the sequence contribution. More potent
ion-pairing systems such as Triethylammonium ion with Hexafluo-
roisopropanol counter ion provide for more regular “charge-based”
separations (Figure 4).
Figure 4: Impact of Ion-pairing System on Separation of a 10-30mer
Heterooligonucleotide Ladder
HPLC system:
Waters BioAlliance
™
2796, PDA Detector with micro UV cell
Sample:
20 mer: TCC CTA GCG TTG AAT TGT CC
25 mer: TCC CTA GCG TTG AAT TGT CCC TTA G
30 mer: TCC CTA GCG TTG AAT TGT CCC TTA GCG GGT
Ladder was prepared by hydrolyzing detritylated
20, 25, and 30mer oligonucleotides with a
3’-exonuclease
Column:
Waters XBridge
™
OST C
18
, 2.5 µm (4.6 x 50 mm)
Mobile phases:
Upper chromatogram: 0.1 M TEAA with acetonitrile gradient; Lower
chromatogram: 16.3 mM TEA - 400 mM HFIP with methanol gradient
Flow rate:
1.0 mL/min
Column Temp.:
60 ˚C
Gradient delay:
0.45 mL
Detection:
260 nm, 5 scans per second
ii. saMple preparation
1. Dissolve the detritylated synthetic oligonucleotide sample in Mobile
Phase A (e.g., 0.1 M TEAA). For example, a 0.05 - 0.2 µmole scale
synthesis can be prepared in 0.1 mL of 0.1 M TEAA. Proportionately
larger or smaller volumes of 0.1M TEAA are required when dissolving
samples from different scale syntheses. Due to the nature of gradient
separations, relatively large volumes of sample (in low organic
strength eluent) can be injected and concentrated onto the head of
the column before beginning the gradient elution program.
2. Samples must be completely in solution and free of particulates
before injecting onto the column. Remove all particles from the
sample (Controlled Pore Glass Synthesis Support, etc.), which may
block the inlet column frit, increase the operating pressure, and
shorten the column life time. Sample contamination with high con-
centration of salts and/or detergents may also interfere with analysis.
