The importance of sample preparation, General considerations, Cell lysis – Bio-Rad GS-900™ Calibrated Densitometer User Manual
Page 8
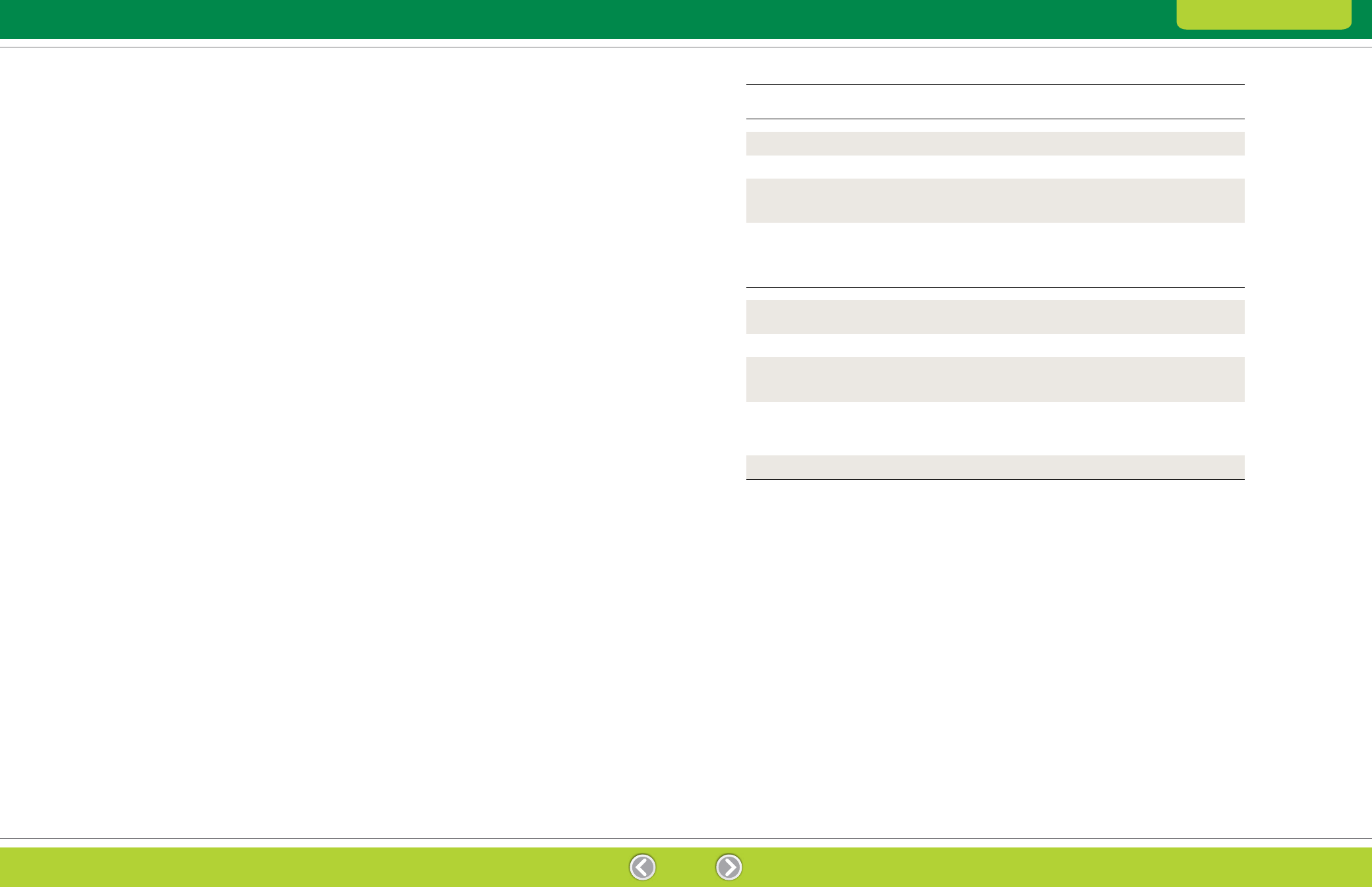
Table 2.1. Suitability of cell disruption methods for various sample types.
Yeast,
Green
Mammalian
Algae, Plant
Soft Cell
Technique
Description
Bacteria Fungi Seeds Material Tissues Culture
Gentle Methods
Osmotic lysis
Suspension of cells in hypotonic solution;
—
—
—
—
—
•
cells swell and burst, releasing cellular contents
Freeze-thaw lysis
Freezing of cells in liquid nitrogen
—
—
—
—
—
•
and subsequent thawing
Detergent lysis
Suspension of cells in detergent-containing
—
—
—
—
—
•
solution to solubilize the cell membrane;
this method is usually followed by another
disruption method, such as sonication
Enzymatic lysis
Suspension of cells in iso-osmotic solutions
•
•
—
•
—
—
containing enzymes that digest the cell wall
(for example, cellulase and pectinase for plant
cells, lyticase for yeast cells, and lysozyme for
bacterial cells); this method is usually followed by
another disruption method, such as sonication
Harsher Methods
Sonication
Disruption of a cell suspension, cooled on ice
•
•
—
—
—
•
to avoid heating and subjected to short bursts
of ultrasonic waves
French press
Application of shear forces by forcing a cell
•
•
—
•
—
•
suspension through a small orifice at high pressure
Grinding
Breaking cells of solid tissues and microorganisms
•
•
•
•
•
—
with a mortar and pestle; usually, the mortar is
chilled with liquid nitrogen and the tissue or cells
are ground to a fine powder
Mechanical
Homogenization with either a handheld device
—
—
—
•
•
—
homogenization
(for example, Dounce and Potter-Elvehjem
homogenizers), blenders, or other motorized
devices; this approach is best suited for soft,
solid tissues
Glass-bead
Application of gentle abrasion by vortexing
•
•
—
—
—
•
homogenization
cells with glass beads
12
13
2-D Electrophoresis Guide
Theory and Product Selection
Chapter 2: Sample Preparation
■
■
Solubilize proteins in a solution that is compatible
with IEF. Incubate proteins in 2-D lysis solution for
at least 30 min at room temperature (denaturation,
solubilization, and disaggregation are time-
dependent processes)
■
■
Determine the amount of total protein in each
sample using a protein assay that is compatible
with chemicals in your samples
■
■
Avoid freeze-thaw cycles; use protein extracts
immediately or aliquot them into appropriately sized
batches and store them at –70°C
Cell Lysis
The effectiveness of a cell lysis method determines
the accessibility of intracellular proteins for extraction
and solubilization. Different biological materials require
different lysis strategies, which can be divided into two
main categories: gentle methods and harsher methods
(Table 2.1).
■
■
Use gentle cell disruption protocols with cells that
lyse easily, such as blood cells and tissue culture cells
■
■
Use harsher methods, which are based mainly on
mechanical rupture (Goldberg 2008), with biological
materials that have tough cell walls (for example,
plant cells and tissues, and some microbes)
■
■
When working with a new sample, compare at least
two different cell disruption protocols with respect to
yield (by protein assay) and qualitative protein content
(by one-dimensional SDS-PAGE)
■
■
Optimize the power settings of mechanical rupture
systems and the incubation times of lysis approaches
■
■
Mechanical cell lysis usually generates heat;
use cooling where required to avoid overheating
the sample
A number of other components are often added to
disruption protocols. Sand, resin, or glass beads
facilitate the disruption of tissues and of plant and
yeast cell walls when added to manual grinding
procedures. Hypotonic buffers cause cells to burst
more readily under physical shearing, and enzymes
such as cellulase, pectinase, lyticase, and lysozyme
are added to break down plant, yeast, and bacterial
cell walls. Nucleases can be added to remove nucleic
acids, which can increase sample viscosity and
interfere with subsequent separation (see the Removal
of Interfering Substances section).
The Importance of Sample Preparation
Sample preparation contributes significantly to
the overall reproducibility and accuracy of protein
expression analysis (Link 1999, Rabilloud 1999,
Molloy 2000). Without proper sample preparation,
proteins may not separate from one another or may
not be represented in the 2-D pattern.
A successful sample preparation strategy enhances
separation quality by:
■
■
Effectively and reproducibly solubilizing proteins
of interest
■
■
Preventing protein aggregation and loss of solubility
during IEF
■
■
Preventing proteolysis or other chemical or
enzymatic protein modifications
■
■
Removing or minimizing the effect of contaminants
such as salts, detergents, nucleic acids, and other
interfering molecules
■
■
Yielding proteins of interest at detectable levels,
which may require fractionation to reduce protein
sample complexity or removal of interfering abundant
or irrelevant proteins
This chapter provides an overview of the principles and
recent developments in sample preparation strategies
prior to first-dimension IEF.
General Considerations
Since protein types and sample origins show great
diversity, there is no universal sample preparation
method. In addition, some proteins simply cannot
be solubilized under conditions compatible with IEF.
Sample preparation procedures must be optimized
empirically and tailored to each sample type and
experimental goal. The following general sample
preparation guidelines should be kept in mind:
■
■
Keep the sample preparation workflow as simple
as possible; increasing the number of sample
handling steps may increase variability and the
risk of sample loss
■
■
With cell or tissue lysates, include protease inhibitors
to minimize artifacts generated by proteolysis;
protease inhibitors are generally not required for
samples like serum or plasma
All but the most gentle cell disruption methods destroy
the compartmentalization of a cell, causing the
release of hydrolases (phosphatases, glycosidases,
and proteases). These enzymes modify proteins in
the lysate, which complicates differential analysis.
The data generated by 2-D electrophoresis are only
meaningful when the integrity of the sample proteins
reflects the state in which they are found in the living
organism. Avoid enzymatic degradation by using one
or a combination of the following techniques:
■
■
Disrupt the sample or place freshly lysed samples in
solutions containing strong denaturing agents such
as 7–9 M urea, 2 M thiourea, or 2% SDS. In this
environment, enzymatic activity is often negligible
■
■
Perform cell lysis at low temperatures to diminish
enzymatic activity
■
■
Lyse samples at pH >9 by adding a base such
as sodium carbonate or Tris(hydroxymethyl)-
aminomethane (Tris) to the lysis solution
(proteases are often least active at basic pH)
■
■
Add protease inhibitors to the lysis solution.
Examples include either small molecules,
such as phenylmethylsulfonyl fluoride (PMSF),
aminoethyl-benzene sulphonyl fluoride (AEBSF),
tosyl lysine chloromethyl ketone (TLCK),
tosyl phenyl chloromethyl ketone (TPCK),
ethylenediaminetetraacetic acid (EDTA), and
benzamidine, or peptide protease inhibitors such
as leupeptin, pepstatin, aprotinin, and bestatin.
For best results, use a combination of inhibitors
in a protease inhibitor cocktail
■
■
If protein phosphorylation is to be studied, include
phosphatase inhibitors such as fluoride or vanadate
Following cell disruption:
■
■
Check the efficacy of cell disruption by light
microscopy (if the sample is a cell suspension)
■
■
Centrifuge all extracts extensively (20,000 × g for
15 min at 15°C) to remove any insoluble material;
solid particles may block the pores of the IPG strip
