Transition from first to second dimension, Molecular weight estimation, Power conditions and reagents for sds-page – Bio-Rad GS-900™ Calibrated Densitometer User Manual
Page 28: Casting sds-page gels using multi-casting chambers, Molecular weight (mw) estimation
52
53
2-D Electrophoresis Guide
Theory and Product Selection
Chapter 4: The Second Dimension: SDS-PAGE
Transition from First to Second Dimension
The transition from first- to second-dimension gel
electrophoresis involves the following:
■
■
Equilibration, which involves two steps that treat
the focused IPG strips with an SDS-containing
buffer to prepare the proteins for SDS-PAGE.
The first equilibration solution contains buffer,
urea, glycerol, reductant, SDS, and dye (optional).
The second equilibration step replaces the
reductant with iodoacetamide to alkylate the
thiol groups. Equilibration ensures the proteins
are coated with dodecyl sulfate and all cysteines
are reduced and alkylated
■
■
Embedding of the strip on the top of the second-
dimension gel. The equilibrated IPG strips are placed
on top of the gel and sealed in place with molten
agarose solution to ensure good contact between
the gel and the IPG strip
Methods for equilibrating and embedding IPG strips
onto second-dimension gels are available in Part II
of this guide.
Power Conditions and Reagents
for SDS-PAGE
For SDS-PAGE, use running conditions that provide
optimum separation across the size range of interest
and that maintain the temperature of the system
during operation. For a complete discussion of running
conditions and the parameters that affect them, please
refer to A Guide to Polyacrylamide Gel Electrophoresis
and Detection, Bio-Rad bulletin 6040. For second-
dimension SDS-PAGE, include a short, low voltage
(50 V) step at the beginning of the run to ensure that all
of the proteins are removed from the IPG strip before
final voltages are applied.
Casting SDS-PAGE Gels Using
Multi-Casting Chambers
In general, proteomics work requires running
several IPG strips and second-dimension gels
per experiment. It is important that gels have a
very similar composition. The best way to ensure
that handcast gels have the same composition is
to cast them at the same time in a multi-casting
chamber. This is especially important when casting
gradient gels. Details of the assembly and use
of multi-casting chambers are available in their
accompanying instruction manuals. Tips that
generally apply to all multi-casting systems are:
■
■
Before assembling the casting chamber, glass
plates should be carefully cleaned with Bio-Rad
cleaning concentrate and thoroughly rinsed with
deionized water
■
■
Each pair of glass plates (two per gel) should
be separated from the next by a spacer sheet;
the spacer sheet allows easier separation of the
cassettes after gel polymerization
■
■
The volume of gel solution should be determined
by measuring the volume of water needed to fill the
assembled glass plates to the desired level in the
multi-casting stand
■
■
Allow overnight polymerization to compensate for
the low concentrations of catalysts (recommended
to ensure that polymerization does not start while
the gradient gels are being cast)
Apparatus for casting multiple gels. Multi-casting chambers for 12 PROTEAN Plus
™
gels or for 12 Mini-PROTEAN gels allow uniform casting
of gradient gels. Gradient makers are available for both size formats.
Molecular Weight (MW) Estimation
SDS-PAGE is a reliable method for estimating the
MW of an unknown protein. The migration rate of a
protein–SDS complex is inversely proportional to the
logarithm of its MW: the larger the polypeptide, the
more slowly it migrates in a gel. The key to accurate
MW determination is selecting separation conditions
that produce a linear relationship between log (MW)
and migration within the likely MW range of the
unknown protein. These parameters are discussed
more thoroughly in Molecular Weight Determination
by SDS-PAGE (bulletin 3133), and a protocol for MW
estimation is provided in Part II of this guide.
For best results, separate the protein sample
on the same gel with a set of protein standards.
See The Little Book of Standards (bulletin 2414) and
the Protein Standards Application Guide (bulletin 2998)
for more information regarding selection of protein
standards. Mixtures of standard proteins with known
MW can be unstained, prestained, or include tags
for development with various secondary reagents
(useful when blotting). Standards can be run in a
reference well or attached to the end of a focused IPG
strip by filter paper onto the second-dimension gel.
For convenience, Bio-Rad’s Precision Plus Protein
standard plugs (catalog #161-0378), which are
embedded in agarose plugs, can also be used.
After separation, determine the relative migration
distance (R
f
) of the protein standards and of the
unknown protein. R
f
is defined as the mobility of
a protein divided by the mobility of the ion front
(Figure 1). Because the ion front can be difficult to
locate, mobilities are normalized to the tracking dye
that migrates only slightly behind the ion front:
R
f
= (distance to band)/(distance to dye front)
Using the values obtained for the protein standards,
plot a graph of log (MW) vs. R
f
(see below). The plot
should be linear for most proteins, provided they
are fully denatured and that the gel percentage
is appropriate for the MW range of the sample.
The standard curve is sigmoid at extreme MW values,
because the sieving affect of the matrix is so large at
high MW that molecules are unable to penetrate the
gel; but at low MW, the sieving effect is negligible and
proteins migrate almost freely. To determine the MW
of the unknown protein band, interpolate the value
from this graph (Figure 2).
Gradient SDS-PAGE gels can also be used to estimate
MW. In this case, log (MW) is proportional to log
(%T). With linear gradients, %T is proportional to the
distance migrated, so the data can be plotted as log
(MW) vs. log (migration distance). Standard curves are
actually sigmoid. The apparent linearity of a standard
curve may not cover the full MW range for a given
protein mixture in a particular gel. However, log (MW)
varies sufficiently slowly to allow fairly accurate MW
estimates to be made by interpolation, and even
extrapolation, over relatively wide ranges.
3.0
2.0
1.0
0
0
0.2
0.4
0.6
0.8
1.0
R
f
lo
g M
W
y = –1.9944x + 2.7824
r
2
= 0.997
Standards
Unknown
Fig. 2. Determining the MW of an unknown protein by SDS-PAGE.
A standard curve of the log (MW) versus R
f
was generated using the
Precision Plus Protein standards from Figure 1. The strong linear
relationship (r
2
> 0.99) between the proteins’ MW and migration
distance demonstrates exceptional reliability in predicting MW.
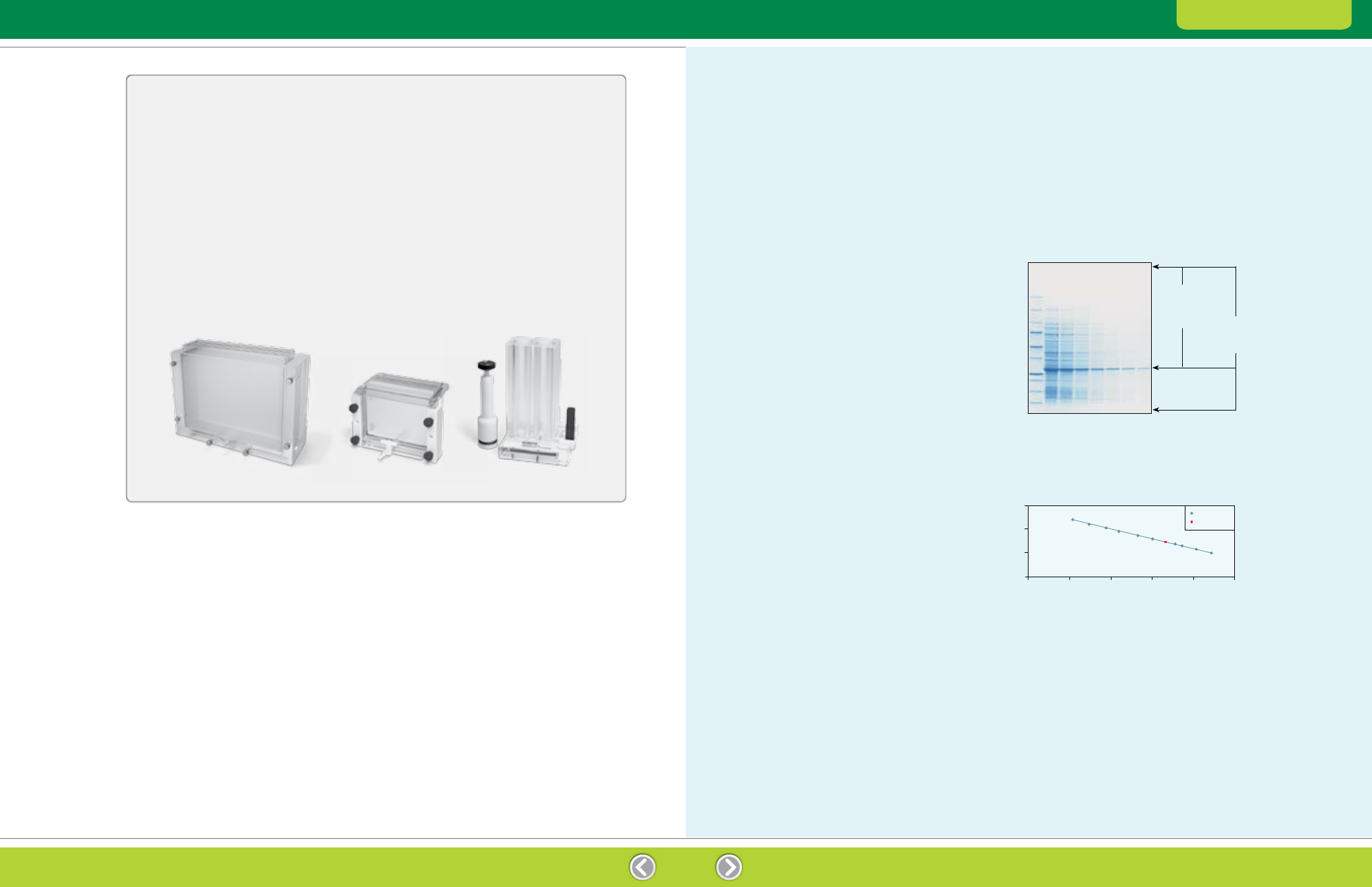
Fig. 1. Example showing MW determination of an unknown
protein. Lane 1, 10 μl of Precision Plus Protein unstained standards;
lanes 2–8, a dilution series of an E. coli lysate containing a
hypothetical unknown protein (GFP). Proteins were separated by
SDS-PAGE in a Criterion 4–20% Tris-HCI gel and stained with
Bio-Safe Coomassie stain.
MW, kD
250
150
100
75
50
37
25
20
15
10
1 2 3 4 5 6 7 8
Top of resolving gel
Migration
distance
of dye front
(67 mm)
Unknown band
Migration
distance of
unknown
band
(45 mm)
Dye front
