Sample quantitation (protein assays), Quantitation of dna, rna, and oligonucleotides, Monitoring bacterial culture growth – Bio-Rad GS-900™ Calibrated Densitometer User Manual
Page 17: Simple kinetic assays, Protein labeling techniques, Fractionation using electrophoresis methods, Produced by the peptide-mediated reduction of cu, Generated by peptide- mediated reduction of cu
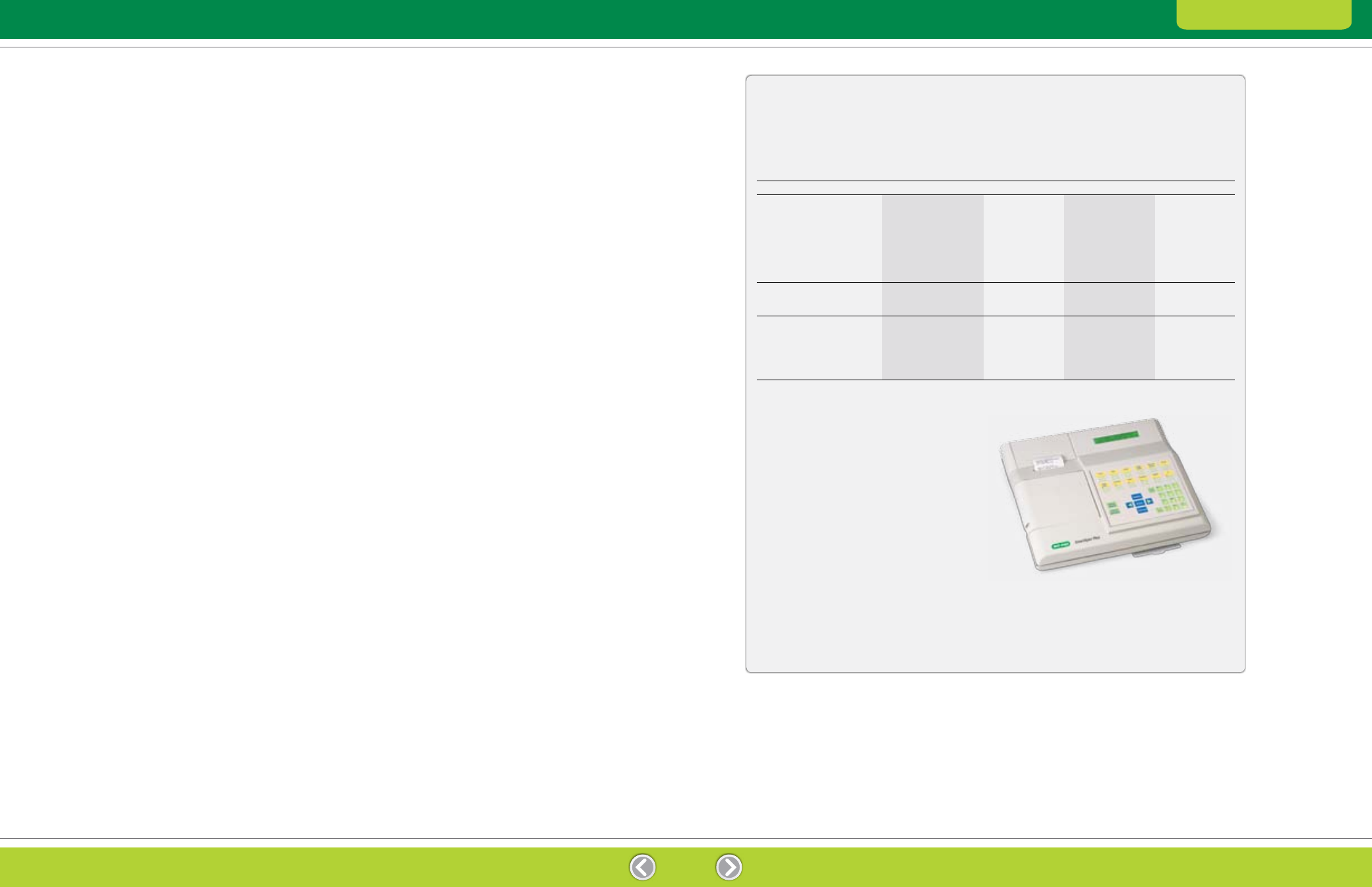
Table 2.3. Bio-Rad protein assay selection guide.
Quick Start
™
Bradford
Bio-Rad
DC
™
RC DC
Method
Bradford
•
•
—
—
Lowry
—
—
•
•
Description
One-step determination;
Standard Bradford
Detergent compatible
Reducing agent
not for use with
assay, not to be
(DC); Lowry assay
and detergent
SDS-containing samples
used with elevated
modified to save time
compatible (RC DC)
levels of detergents
and to be more
(>0.1% SDS)
accurate
Standard-concentration Assay
Sample volume
100 µl
100 µl
100 µl
100 µl
Linear range
0.125–1.5 mg/ml
0.125–1.5 mg/ml
0.125–1.5 mg/ml
0.2–1.5 mg/ml
Low-concentration Assay
Sample volume
1 ml
800 µl
200 µl
200 µl
Linear range
1.25–25 µg/ml
1.25–25 µg/ml
5–250 µg/ml
5–250 µg/ml
Microplate assay volume
5 µl
10 µl
5 µl
**
Minimum incubation
5 min
5 min
15 min
15 min
Assay wavelength
595 nm
595 nm
650–750 nm
650–750 nm
Protein Assay Products
SmartSpec Plus Spectrophotometer
Protein concentration in 2-D sample solutions
is best measured using the RC DC
™
protein
assay, a modification of the Lowry assay that
incorporates a precipitation step that removes
SmartSpec Plus Spectrophotometer
reducing agents and detergents. For more
information on protein quantitation using visible
assays, refer to Bio-Rad bulletin 1069.
The color change observed in protein assays is
measured using a spectrophotometer. Bio-Rad’s
SmartSpec Plus spectrophotometer has
preprogrammed methods for protein quantitation
and a working wavelength range of 200–800 nm.
It can be used for routine applications such as:
■
■
Quantitation of proteins via the Bradford,
Lowry, and BCA assay methods
■
■
Quantitation of DNA, RNA, and oligonucleotides
■
■
Monitoring bacterial culture growth
■
■
Simple kinetic assays
■
■
Wavelength scans with peak detection
Features built into the SmartSpec assay
methods facilitate data collection and present
a complete analysis of assay results. Bio-Rad
also offers compatible quartz and UV-transparent
plastic cuvettes.
30
31
2-D Electrophoresis Guide
Theory and Product Selection
Chapter 2: Sample Preparation
Additional Resources
Samples can be prepared for 2-D electrophoresis
using many other techniques. Consult Posch (2008)
for more information on:
■
■
Sample preparation basics (cell disruption, sample
solubilization, protein assays, contaminant removal)
■
■
Protein labeling techniques
■
■
Fractionation using chemical reagents
and chromatography
■
■
Fractionation using electrophoresis methods
■
■
Enrichment strategies for organelles, multiprotein
complexes, and specific protein classes
■
■
Application of sample preparation tools and
fractionation strategies to study different
biological systems
Sample Quantitation (Protein Assays)
Determine the concentration of protein in a sample
(Berkelman 2008) by protein assay to:
■
■
Ensure that the amount of protein to be
separated is appropriate for the IPG strip length
and visualization method
■
■
Facilitate comparison among similar samples;
image-based analysis is simplified when equivalent
quantities of proteins have been separated
The most commonly used protein assays are
visible assays, assays in which the presence of protein
causes a visible color change that can be measured
with a spectrophotometer (Sapan et al. 1999;
Noble and Bailey 2009; see the Protein Assay
Products and SmartSpec
™
Plus Spectrophotometer
sidebar). All protein assays utilize a dilution series of a
known protein (usually bovine serum albumin or bovine
g-globulin) to create a standard curve from which the
concentration of the sample is derived (for a protocol
describing protein quantitation, refer to Part II of
this guide).
The chemical components of the sample buffer and
the amount of protein available for assay dictate the
type of assay that may be used.
■
■
Bradford assays (Bradford 1976) — based on an
absorbance shift of Coomassie (Brilliant) Blue G-250
dye under acid conditions, when a redder form of
the dye is converted into a bluer form upon binding
to protein. The increase of absorbance at 595 nm
is proportional to the amount of bound dye and,
therefore, to the amount (concentration) of protein
present in the sample. In comparison to other protein
assays, the Bradford protein assay is less susceptible
to interference by various chemicals that may be
present in protein samples, with the exception of
elevated concentrations of detergents like SDS.
The response of the Bradford protein assay is
only slightly affected by urea, thiourea, and
CHAPS in concentrations up to 1.75 M, 0.5 M,
and 1% (w/v), respectively
■
■
Lowry (Lowry et al. 1951) — combines the
reactions of cupric ions with the peptide bonds
under alkaline conditions with the oxidation of
aromatic protein residues. The Lowry method is
based on the reaction of Cu
+
, produced by the
peptide-mediated reduction of Cu
2+
, with Folin-
Ciocalteu reagent (a mixture of phosphotungstic acid
and phosphomolybdic acid in the Folin-Ciocalteu
reaction). The Lowry assay is intolerant of thiourea,
reductants such as DTT, and chelating agents
such as EDTA
■
■
BCA (bicinchoninic acid, Smith et al. 1985) —
BCA reacts directly with Cu
+
(generated by peptide-
mediated reduction of Cu
2+
) to produce a purple end
product. The reagent is fairly stable under alkaline
conditions and can be included in the copper
solution to allow a one-step procedure. Like the
Lowry assay, the BCA assay is intolerant of thiourea,
reductants such as DTT, and chelating agents such
as EDTA
2-D sample solutions typically contain reagents
that interfere with all of the assays described above.
The Bradford assay may be used on samples that
are concentrated enough to be diluted with water
so that urea, thiourea, and CHAPS are no longer
present at interfering levels (typically at least fourfold).
Otherwise, modified assay procedures may need to
be employed (see the Protein Assay Products sidebar).
