5 operation, 3 direct photometric measurement of protein – Eppendorf BioPhotometer User Manual
Page 19
61
5 Operation
5.3 Direct photometric measurement of protein
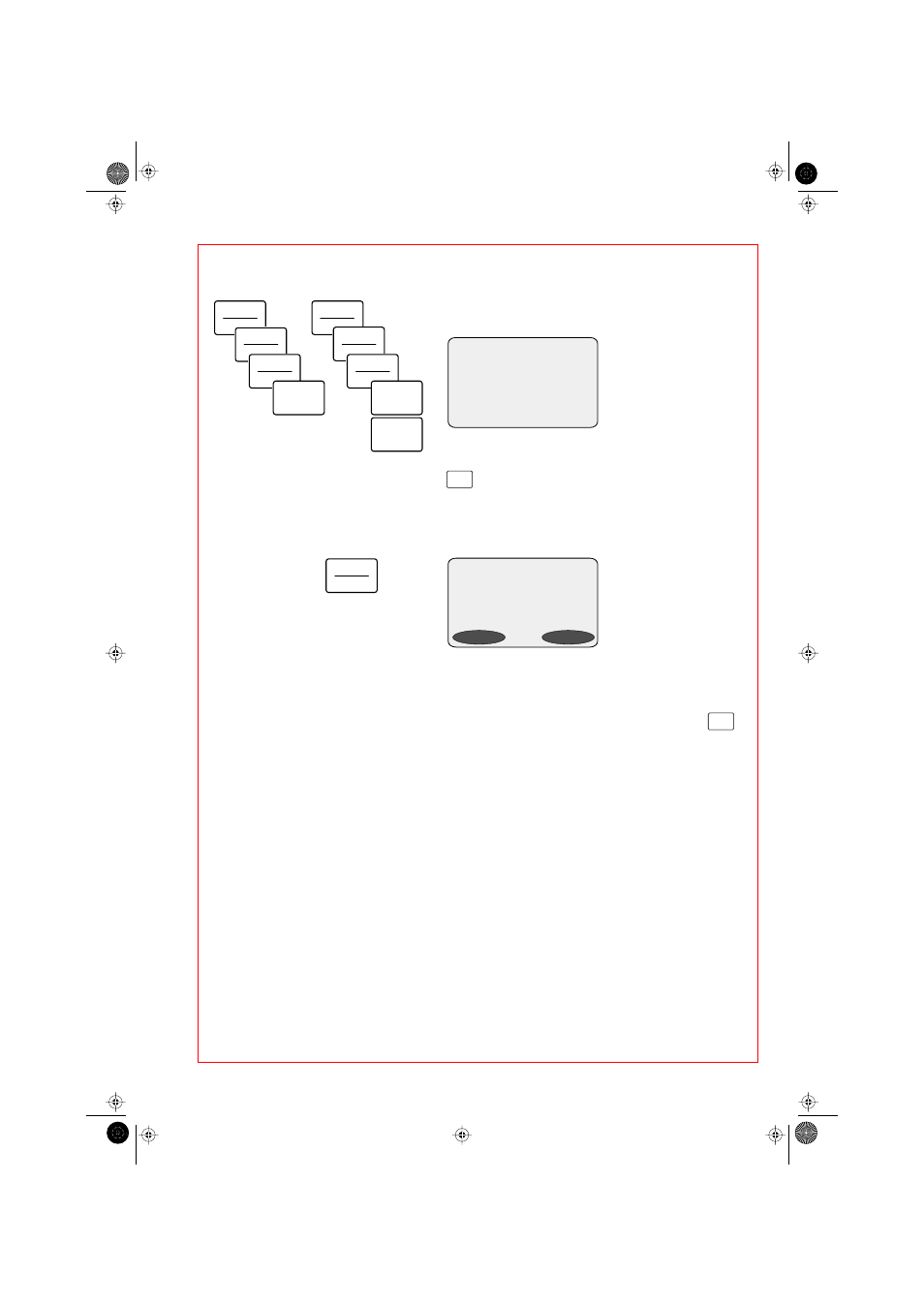
Display after entry of "140
µ
L sample volume" and "300 base
pairs":
The molar unit of concentration (here: "pmol/mL") is
preprogrammed, but can be selected and changed using the
key.
Call up method
Calculation
For the "protein" method, the "absorbance" calculation mode
is stored, i.e. only the absorbances which are measured
directly appear in the display. Calculations via the following
calculation procedures can be programmed using the
key (see Section 6 "Programming"):
– Factor
– Standard (One-point calibration)
– Warburg formula
The number of decimal places of the preprogrammed factor
or the preprogrammed nominal concentration of the standard
determines the number of decimal places in the result.
When programming the factor, please ensure that the factor
is adapted in line with the unit of concentration selected.
Measuring solutions with absorbances lower than app. 0.02
to 0.03 A
280
corresponds to a DNA concentration of app. 1.0
to 1.5
µ
g / m L should not be used. This is because with such
low absorbances, disturbances such as small particles,
microbubbles or turbidity have a great deal of influence upon
the measuring result and often lead to unreliable results.
1
Bradford
4
Protein
0
Sample No.
Enter
3
BCA
0
Sample No.
0
Sample No.
Enter
Enter
d s D N A
S A M P L E 0 0 1
9 . 8
µg
3 5 3 . 5 p m o l / m L
4 9 . 5 p m o l
7
0
.
0
µg/mL
Parameter
4
Protein
P ROT E I N
A B S O R B A N C E
Blank
Sample
or
Parameter
05_Bedienung_e.fm Seite 61 Dienstag, 21. Februar 2006 10:09 Uhr
