BUCHI NIRCal User Manual
Page 149
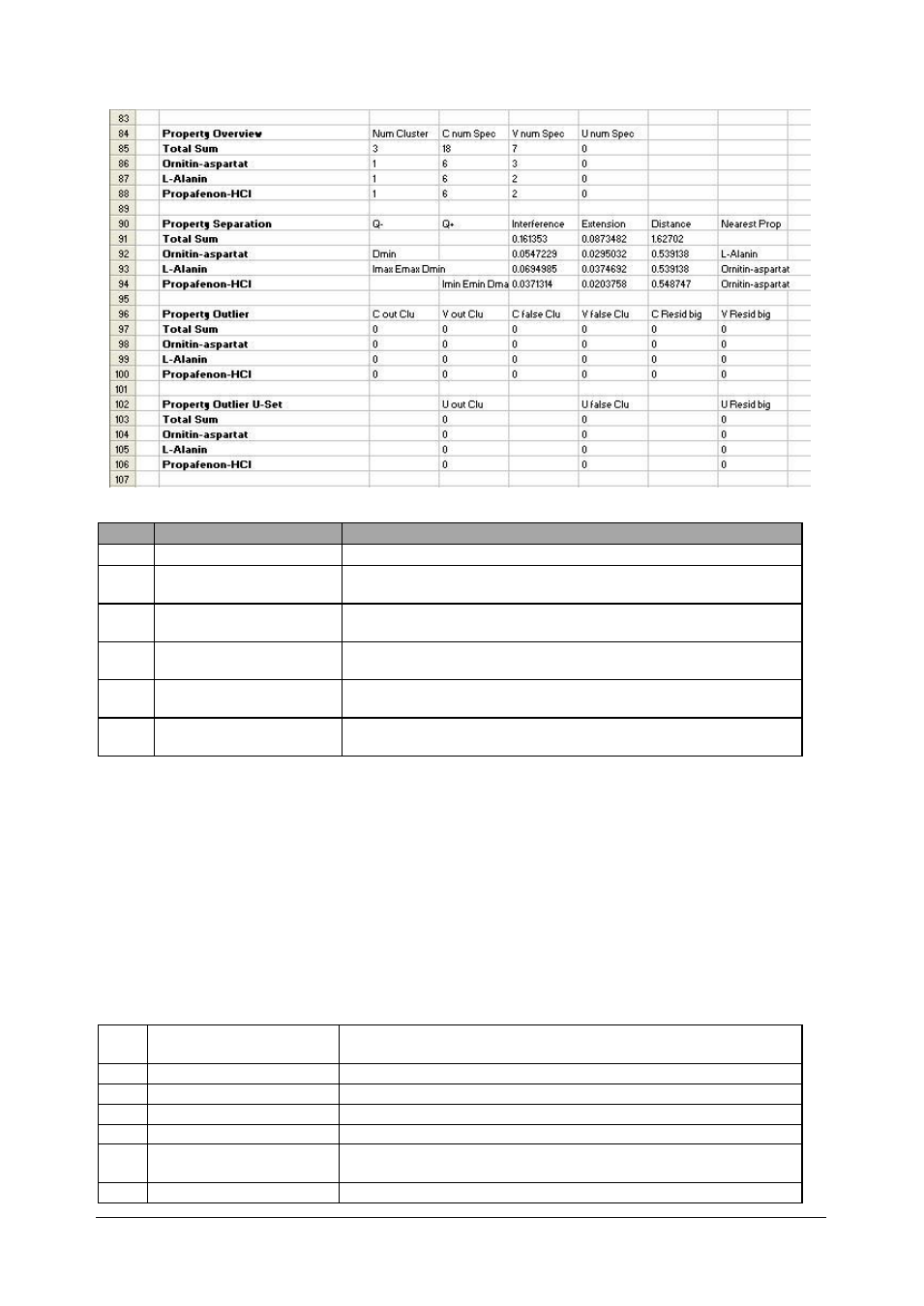
Chemometrics
NIRCal 5.5 Manual, Version A
149
Row
Name
Description
84
Property Overview
Number of clusters and selected spectra
85
Total Sum
Num Cluster = Total number of clusters, should be equal to the
number of calibration properties
86
1st Property
Num Cluster = should be 1, the spreading of C-and V-Set
spectra about 2/3-1/3
87
2nd Property
Num Cluster = should be 1, the spreading of C-and V-Set
spectra about 2/3-1/3
88
3rd Property
Num Cluster = should be 1, the spreading of C-and V-Set
spectra about 2/3-1/3
90
Property Separation
Information about the influence of the properties on the
separation *
* Description of the property separation:
Q- = Imax, Emax and Dmin indicate bad influence on the substance separation
Q+ = Imin, Emin and Dmax indicate good influence on the substance separation
Nearest Prop = Name of the property with the smallest distance
Judging criteria:
Distance = Mahalanobis distance to the closest property (spectrum to spectrum of the other
cluster; see Property Adjacency)
Extension = The max. Mahalanobis distance of each cluster in the factor space (secondary
principal components)
Interference = Extension divided by distance
Imin / Imax = Extreme values regarding the parameter Interference
Emin / Emax = Extreme values regarding the parameter Extension
Dmin / Dmax = Extreme values regarding the parameter Distance
96
Property Outlier
Number of outlier spectra outside a cluster, in false cluster,
residual too big in C- and V-Set
97
Total Sum
All should be 0
98
1st Property
All should be 0
99
2nd Property
All should be 0
100
3rd Property
All should be 0
102
Property Outlier U-Set
Number of outlier spectra outside a cluster, in false cluster,
residual too big in U-Set
103
Total Sum
Normally the U-Set is empty, the sum is also zero
