8 evaluation procedure, Evaluation procedure 8.1, Nucleic acid quantification uv 260 nm with factor – Eppendorf AF2200 Plate Reader User Manual
Page 75
75
Evaluation procedure
Eppendorf
®
PlateReader AF2200
English (EN)
8
Evaluation procedure
8.1
Nucleic acid quantification UV 260 nm with factor
Sample results [Unit] = absorbance * factor sample type * correction factor pathlength
8.2
Nucleic acid and Protein quantification with standards
(Nucleic acid quantification UV 260 nm with standards, Protein quantification UV 280 nm with standards,
Nucleic acid quantification Fluorescence 485/535 nm with standards, Protein quantification BCA 562 (600)
nm with standards, Protein quantification Bradford 595 (600) nm with standards, Protein quantification
Lowry 750 (600) nm with standards)
Sample results are calculated using the curve-fitting model "linear regression". The limit of R
2
is
determined by the user. The user can change the limit for R
2
in the User Settings Dialog of the PlateReader
AF2200 Software.
8.3
Protein quantification Nano Orange 485/595 nm with standards
Sample results are calculated using the curve-fitting model "polynomial 2nd order regression" is used. The
user can change the limit for R
2
in the User Settings Dialog of the PlateReader AF2200 Software.
8.4
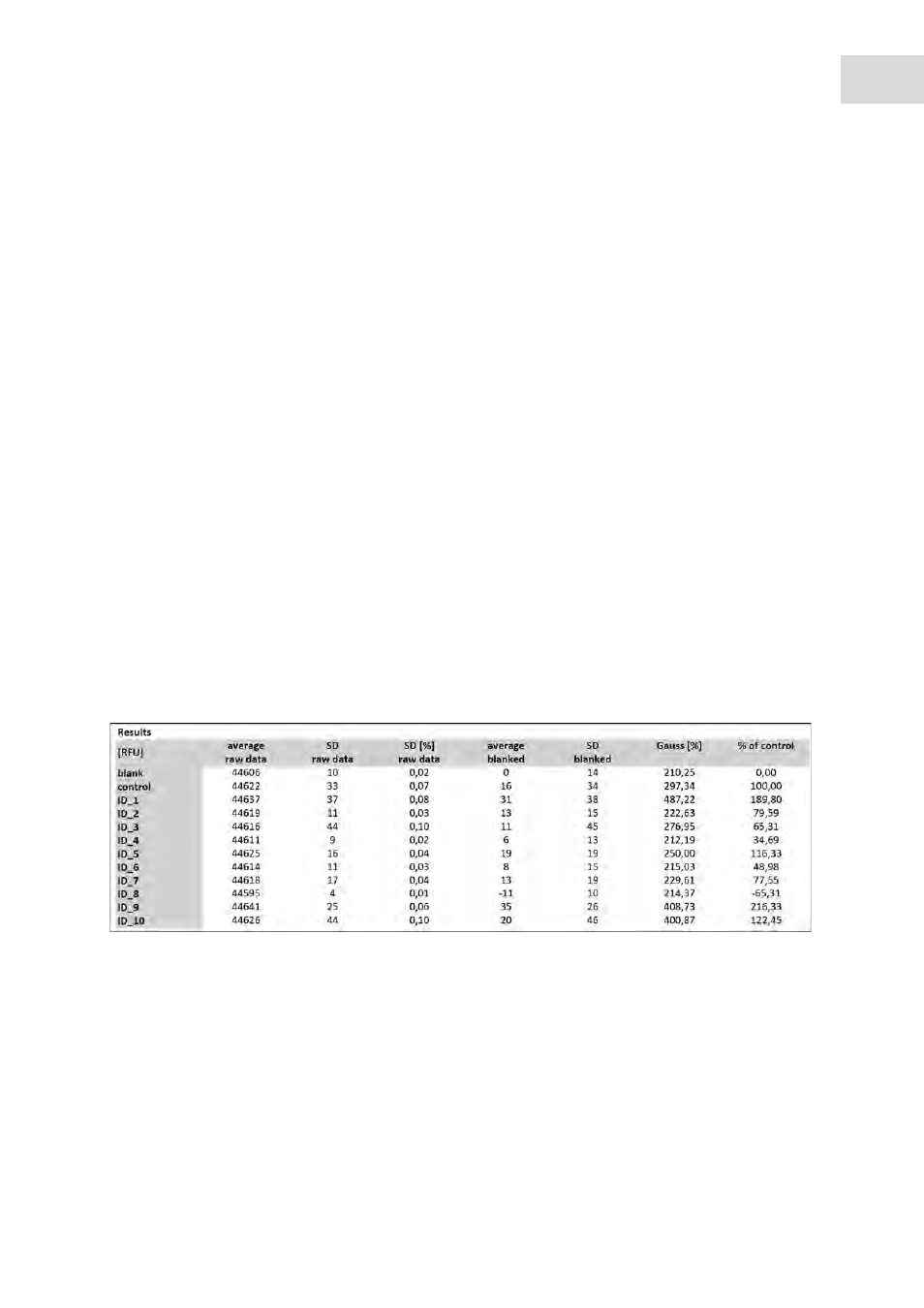
Cell viability CellTiter-Blue Assay, Cell viability Apo-ONE Caspase-3/7 Assay
Abb. 8-1: Results table cell viability
Fig. 8-1:
Results table cell viability
