Bio-Rad CHEF Mapper® XA System User Manual
Page 52
Several agarose types allow easy handling of low concentration gels. These agaroses, in
the concentration range of 0.5–0.8%, can be used to decrease the run time in separation of large
DNA (> 1 mb). An example of this type of agarose is Bio Rad’s Chromosomal Grade Agarose
(catalog number 162-0133).
Buffer Concentration and Temperature
In pulsed field electrophoresis, the mobility of the DNA is sensitive to changes in buffer
temperature. As the buffer temperature increases, the mobility of the DNA increases, but the
band sharpness and resolution decrease. Chill the buffer to 14 °C to maintain band sharpness
and to dissipate heat generated during prolonged runs. Buffer recirculation is required to pre-
vent temperature gradients from occurring. High voltage runs (300 V) exceeding 1 day require
buffer changes after each 24 hour period, to prevent buffer degradation.
Standard tris borate (TBE) at a concentration of 0.5x is the most commonly used buffer
in pulsed field electrophoresis. Tris-acetate buffer (TAE) at a concentration of 1.0x can be
used in place of TBE. Other buffer concentrations used are in the range of 0.25x –1.0x. In
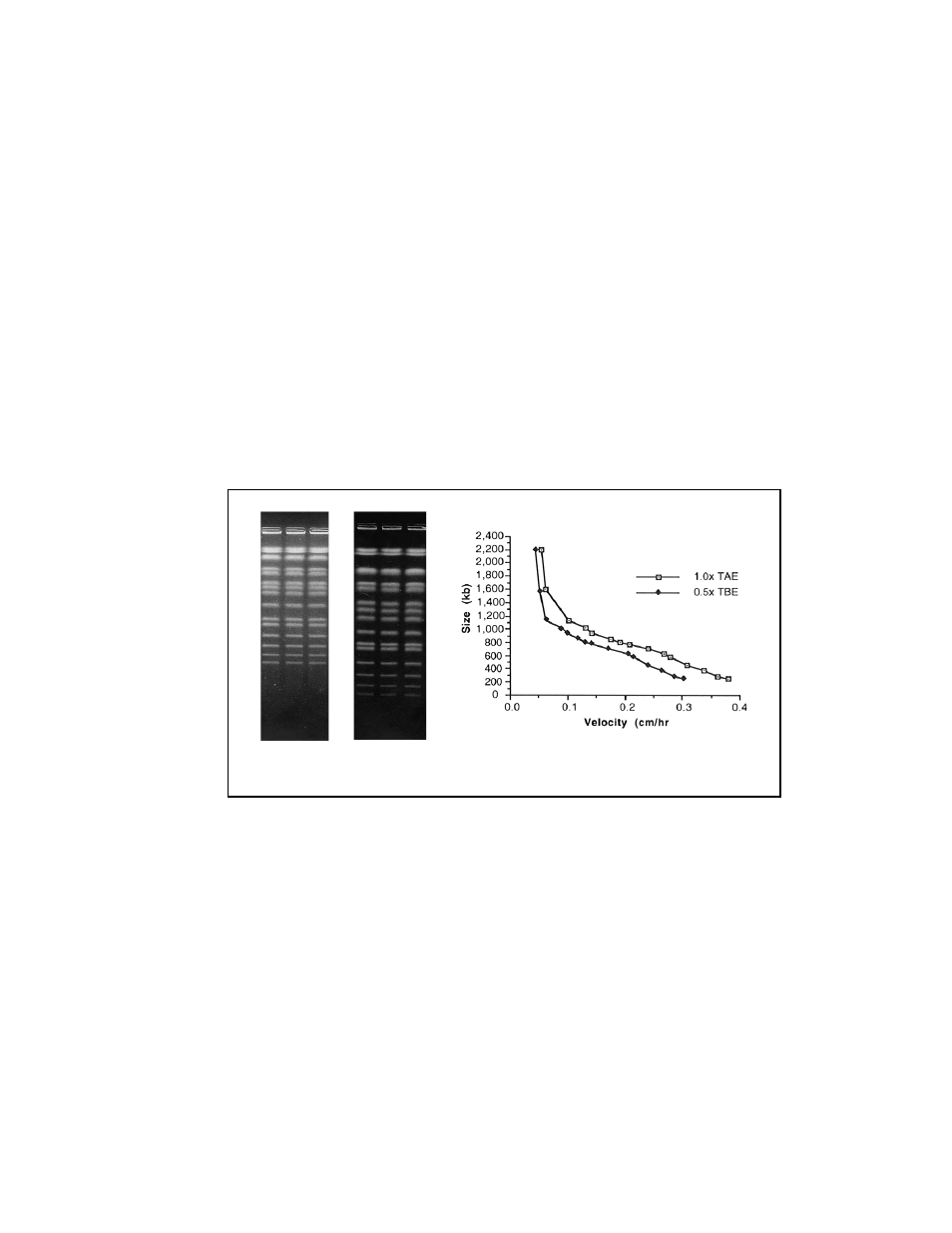
Figure 9.1 two different gels, one using 0.5x TBE and the other using 1.0x TAE, were run to
show the difference in mobility of DNA in the two buffers.
Fig. 9.1. Two gels, one in 0.5x TBE and the other in 1.0x TAE, were run to show the difference in
mobility of DNA in the two buffers.
S. cerevisiae chromosomes were separated on a 1% gel with a
60 second pulse for 15 hours, followed by a 90 second pulse for 9 hours, at 6 V/cm. Notice the
increased migration of the DNA molecules in the TAE gel when compared with the TBE gel.
Switch Times
The migration rate of DNA molecules through an agarose gel depends on pulse time,
voltage (field strength), pulse angle, and run time. In pulsed field electrophoresis, DNA
molecules are subjected to alternating electric fields imposed for a period called the switch
time. Each time the field is switched, the DNA molecules must change direction or re-orient
in the gel matrix. Larger molecules take longer to re-orient and have less time to move during
each pulse, so they migrate slower than smaller molecules. Resolution will be optimal for
DNA molecules with re-orientation times comparable to the pulse time. So, as the DNA size
increases, the pulse time must be increased to resolve the molecules. Under some conditions,
larger molecules may run ahead of smaller ones (Lai, E., Personal Communication).
48
0.5x TBE
1.0x TAE
