Low template input, low expression, high cq values, Setting the baseline and threshold – Bio-Rad SsoAdvanced™ Universal SYBR® Green Supermix User Manual
Page 20
14 |
SsoAdvanced
™
Universal SYBR
®
Green Supermix Instruction Manual
14 |
Low Template Input, Low Expression, High Cq Values
If your Cq values are higher than expected or you are concerned about Cq values >30,
consider the following corrective actions:
1. Confirm the expected expression level, if known, to ensure that the target of interest is
present in your given sample. Additionally, consider higher input concentrations of sample
for low expressing targets. Remember that for every twofold increase in starting sample
concentration, the Cq value shifts one cycle earlier (assuming 100% PCR efficiency).
2. Confirm the template input amount using a fluorescence-based quantification method to
ensure the cDNA input range is 100 ng to 100 fg or the genomic DNA input range is 500 ng
to 5 pg. (cDNA will require purification prior to quantification analysis.)
3. Increase the volume of template pipetted into the PCR reaction. For the highest accuracy
and precision, pipet a minimum volume of 5 µl for each sample.
4. Consider adding a carrier to your sample stock to increase homogeneity — examples
include tRNA, glycogen, and unrelated gDNA.
5. Consider using nonstick polypropylene tubes for sample stock storage to prevent nucleic
acid from binding to the tube walls.
6. Confirm that the reverse transcription reaction was successful. A simple-to-follow protocol is
outlined in
Setting the Baseline and Threshold
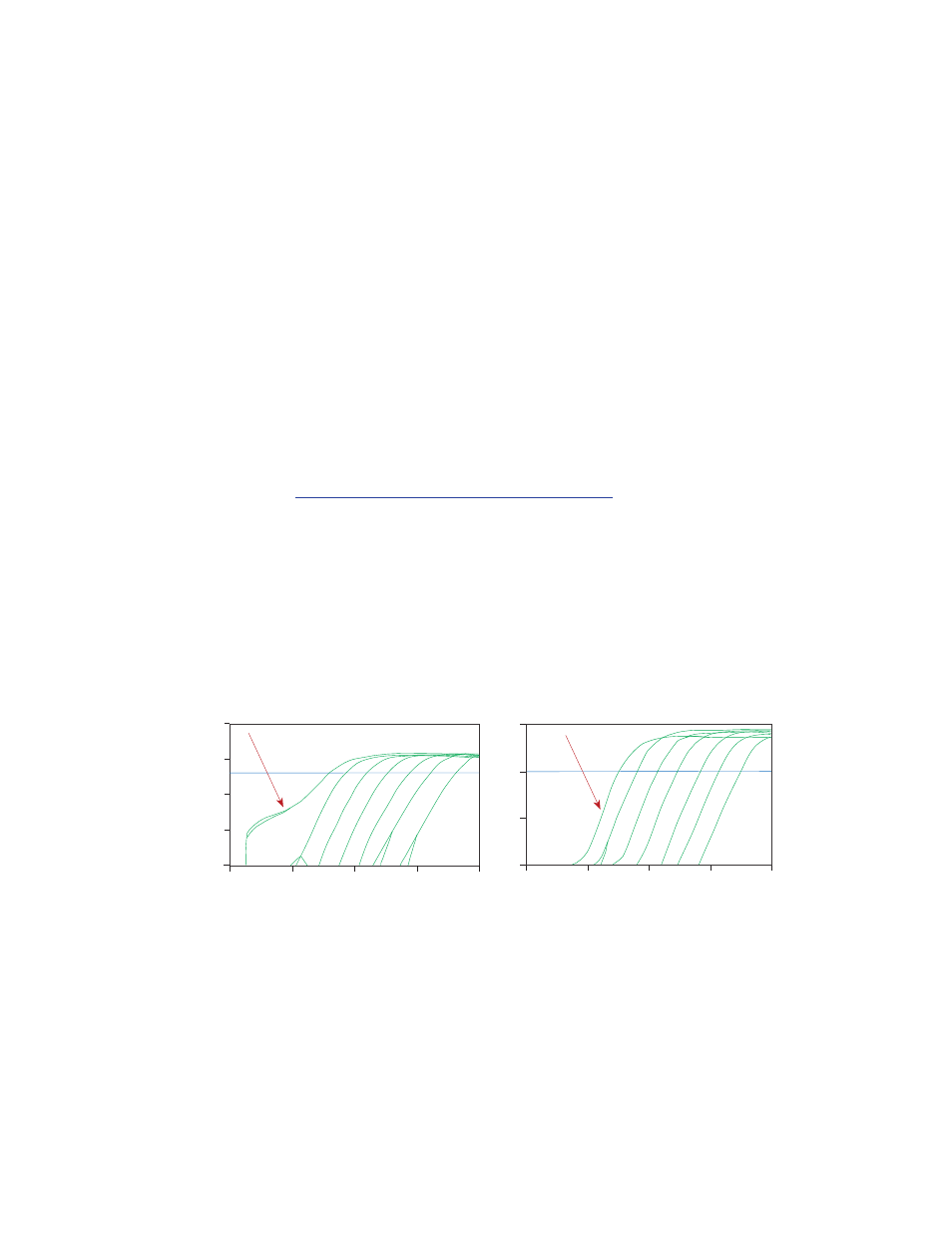
If you notice that any data point(s) in your amplification plots exhibit a sigmoidal shape in
the log view (Figure 10, left), this is typically due to an incorrect baseline setting. Consider
the following corrective actions:
1. Deselect automatic baseline setting and assign manual baseline. Adjust the baseline begin
and end cycles so that the amplification plot matches the others on the plot. Sometimes this
takes a few tries, but a general rule of thumb is to set the end cycle about two cycles before
the start of true amplification, as seen in Figure 11.
Fig. 10. Incorrect baseline is exhibited in the left graph indicated by the arrow pointing to the first
dilution point where the amplification plot is more sigmoidal in shape. As a result, an artificially lower
Cq value is obtained. Corrected baseline is shown in the graph on the right.
R
FU 10
3
10
2
10
1
10
4
10
6
10
4
10
3
10
2
10
1
10
20
30
40
0
0
10
20
30
40
Amplification
Incorrect Baseline
Cycles
Cycles
Correct Baseline
Amplification
R
FU
