Tips for success, Pcr efficiency, Linearity – Bio-Rad SsoAdvanced™ Universal SYBR® Green Supermix User Manual
Page 16
10 |
SsoAdvanced
™
Universal SYBR
®
Green Supermix Instruction Manual
10 |
3. Cycle according to the recommended protocol.
4. Analyze the data. Follow the guidelines in this manual for setting the baseline and threshold
prior to analyzing the data.
Tips for Success
■
■
Pipet a minimum of 5 µl for each sample. This ensures greater precision and a smaller standard
deviation for technical replicates. If the samples are too concentrated, simply dilute accordingly.
Use a calibrated pipet of the appropriate volume range and never plunge the tip more than several
millimeters below the surface of the sample. Pipet slowly and use the pipet tip demarcations to
visualize accuracy
■
■
Prepare individual master mixes for each sample by combining the real-time PCR supermix,
nuclease-free water, and primers along with the template and mix thoroughly. Then, pipet 20 µl
into the respective wells on the plate
■
■
A tenfold dilution series is recommended to cover the most logs of dynamic range; however,
depending on the expression level of the gene(s) evaluated and the total template amount
available, this can be reduced to a fivefold dilution series
PCR Efficiency
Calculate efficiency using the software or the following equation:
E =10
[–1/m]
–1. A PCR efficiency from 90–110% (slope values from –3.6 to –3.1) is preferred.
5. To determine which math model should be applied, simply subtract the slope value of the
reference gene from each target gene. If the ∆slope is ≤0.1, then the PCR efficiencies are
within accepted limits and the ∆∆C
T
math model can be used. If the ∆slope is ≥0.1, then the
efficiency correction math model (Pfaffl method) must be applied.
Linearity
Calculate the R
2
statistic for each standard curve using the qPCR analysis software; the R
2
should be ≥–0.980. However, if the R
2
is <0.980, remove outliers. If there are too many outliers,
then reevaluate the experiment to determine the cause of the lower R
2
value.
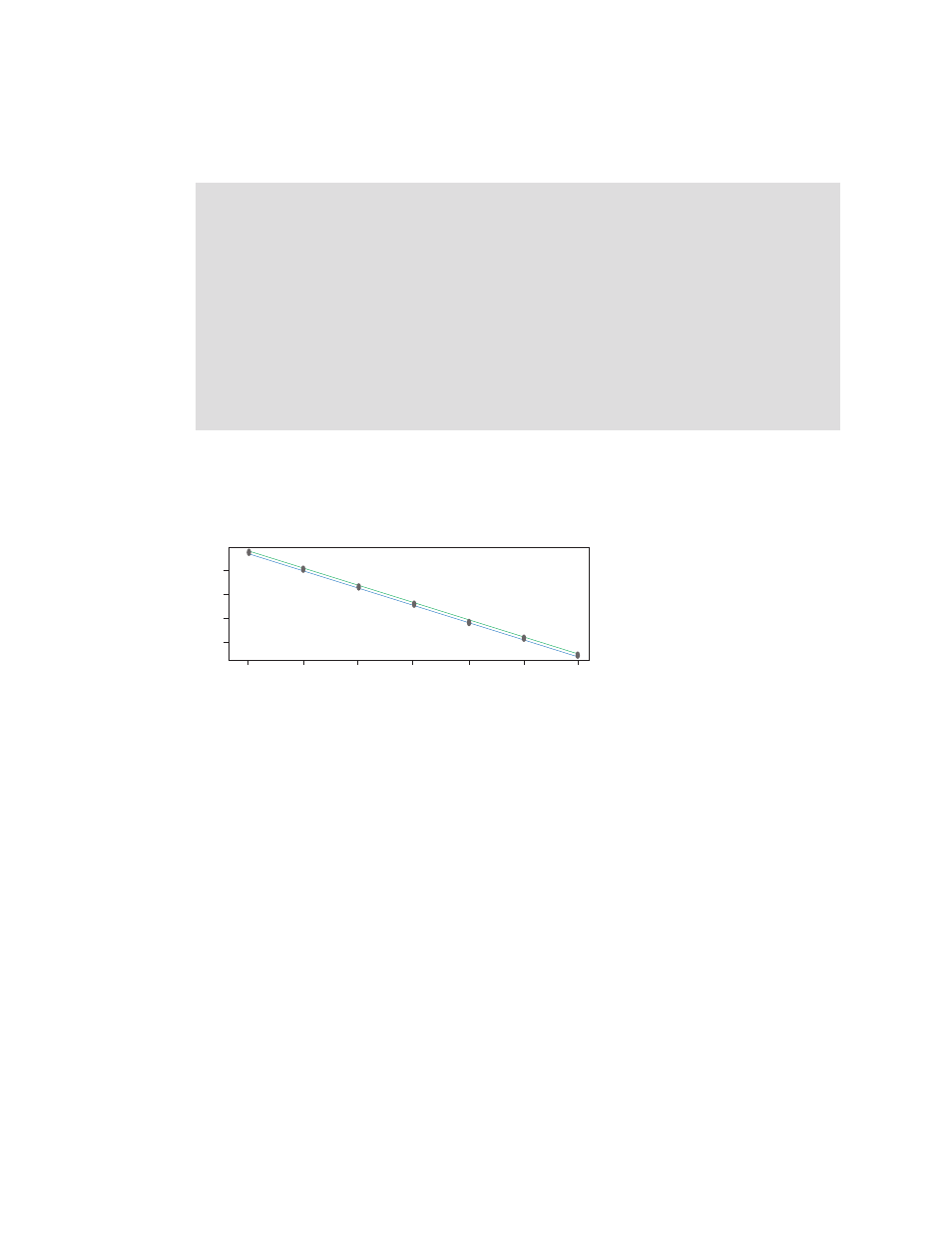
Fig. 7. Reference gene has a PCR efficiency of 97.59% (–3.381) and six
logs of dynamic range. The target gene has a PCR efficiency of 99.17%
(–3.342) with six logs of dynamic range. Subtracting the slope values,
3.381 – 3.342 = 0.039, which is <0.1.
30
2
3
4
5
Log Starting Quantity
Cq
6
7
8
25
20
15
