Determining the optimal reference gene – Bio-Rad SsoAdvanced™ Universal SYBR® Green Supermix User Manual
Page 11
SsoAdvanced
™
Universal SYBR
®
Green Supermix Instruction Manual
| 5
Real-Time PCR Validation for Gene Expression Experiments
The following validation experiments are critical for obtaining valid and publishable real-time
PCR data following the MIQE guidelines. These simple-to-follow experiments should be
completed prior to starting a new real-time PCR project.
Determining the Optimal Reference Gene
To properly perform a gene expression experiment, it is imperative that an optimal
reference gene(s) is used. The reference gene(s) must maintain a consistent expression
level across all samples in the project regardless of treatment, source, or extraction
method. The variation in reference gene expression is somewhat dependent on the level
of fold change discrimination desired. For example, if a twofold change in expression is
important, then the reference gene should have little to no variation in expression. However,
if a 20-fold change in expression is important, then the reference gene expression can have
some variability. To validate a reference gene(s), follow these steps:
1. Begin searching for a candidate list of reference genes by searching publications, speaking
with researchers using similar model systems, and mining microarray data, if available.
Minimally, five reference genes should be selected for evaluation. For your convenience,
Bio-Rad offers pre-plated reference gene panels using our highly validated and optimized
.
2. From your experiment, randomly select a few samples from each group (for example,
treatments, time courses, sources) ensuring that you evaluate all variable sample groups.
3. Isolate the RNA and DNase-treat using the same protocol for all samples. Quantify and
normalize the RNA to the same concentration.
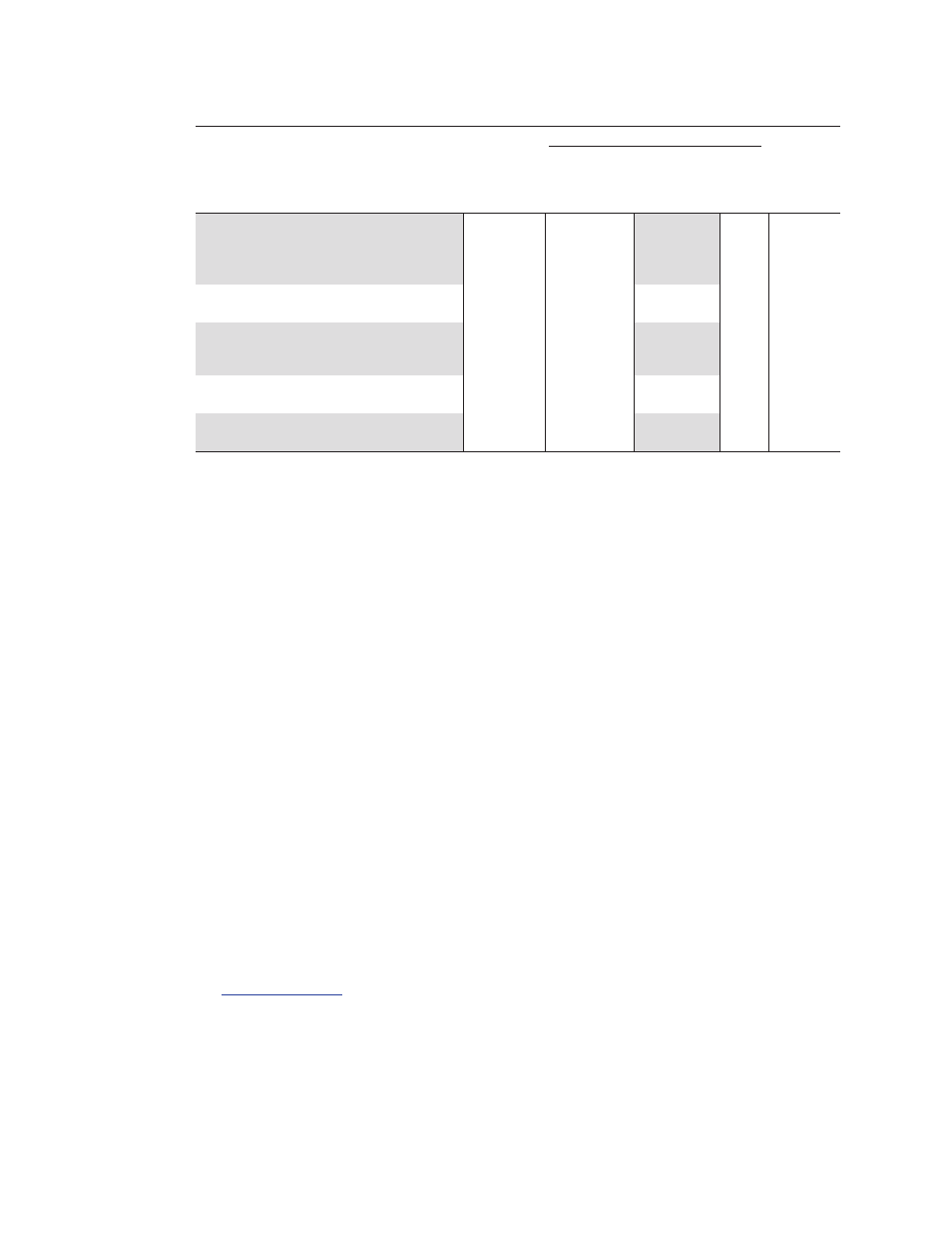
Table 2. Thermal cycling protocol.
Amplification
Polymerase
Annealing/
Activation
Extension +
Setting/
and DNA
Denaturation Plate Read
Melt-Curve
Real-Time PCR System
Mode
Denaturation at 95/98°C
at 60°C** Cycles Analysis
Bio-Rad
®
CFX96
™
,
CFX384
™
, CFX96 Touch
™
,
SYBR
®
only
10–30 sec
CFX384 Touch
™
,
CFX Connect
™
Bio-Rad
®
iQ
™
5, MiniOpticon
™
, Standard
15–30 sec
Chromo4
™
, MyiQ
™
ABI 7500, StepOne,
Fast
5–15 sec
10–30 sec 35–40
StepOnePlus, 7900HT
Standard
60 sec
and ViiA7
Roche LightCycler 480
Fast
10–30 sec
Standard
60 sec
Qiagen Rotor-Gene and
Fast
10–30 sec
Stratagene Mx series
* 98°C is highly recommended for genomic DNA template to ensure complete denaturation.
** Shorter annealing/extension times (1–10 sec) can be used for amplicons <100 bp. Longer annealing/extension
times (30–60 sec) can be used for amplicons >250 bp, GC- or AT- rich targets, crude samples, or for higher input
amounts (for example, 100 ng of cDNA or 50 ng of genomic DNA).
30 sec at
95°C for
cDNA
or
5–15 sec
2–3 min at
98°C for
genomic
DNA*
65–95°C
0.5°C
increment
2–5
35–40 sec/step
(or use
instrument
default
setting)
