Bio-Rad SsoAdvanced™ Universal SYBR® Green Supermix User Manual
Page 14
8 |
SsoAdvanced
™
Universal SYBR
®
Green Supermix Instruction Manual
8 |
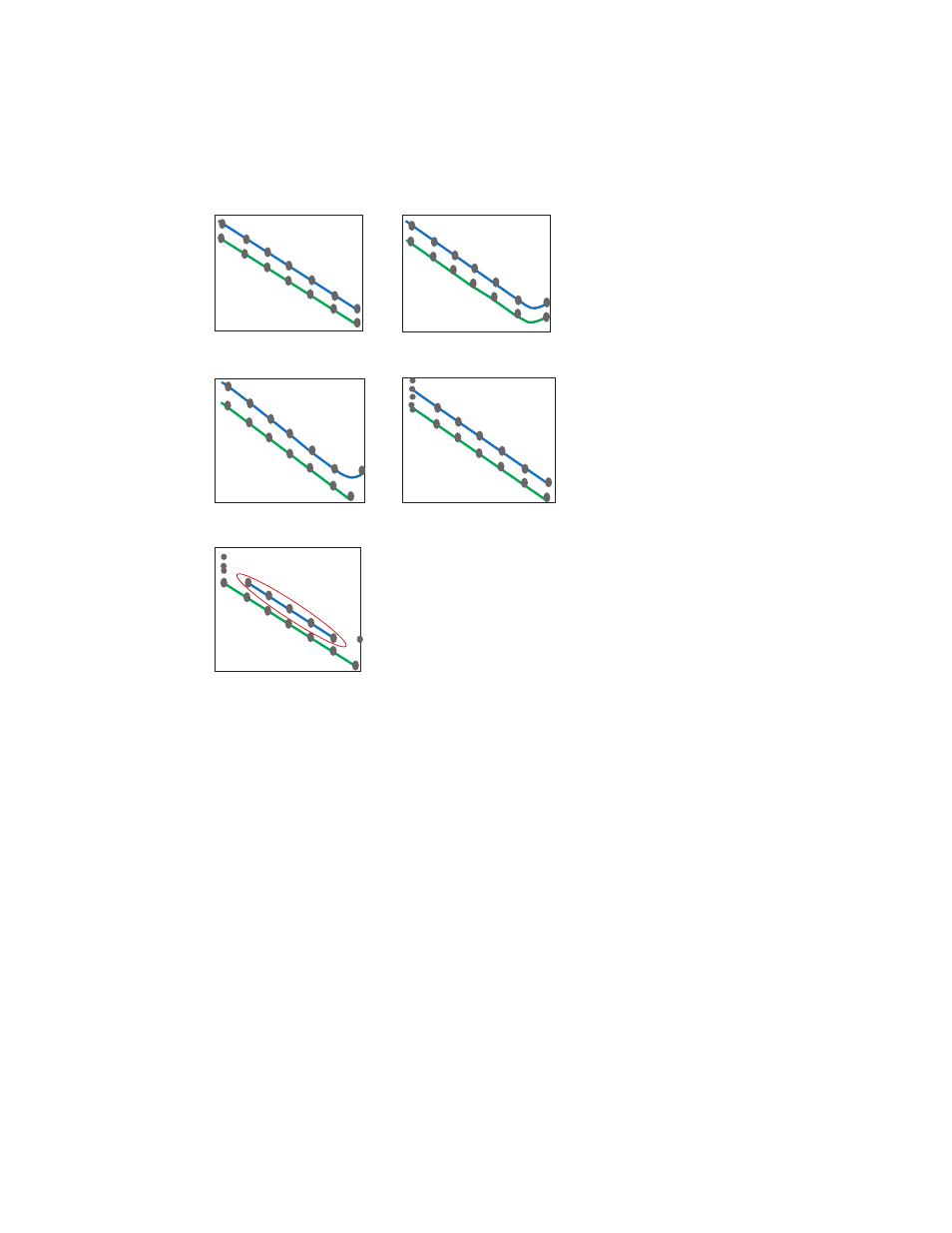
6. Evaluate the data. Follow the guidelines in this manual (page 14–15) for setting the baseline
and threshold prior to analyzing the data. Figure 4 illustrates the most common results from
the experiment and how to interpret the data.
Fig. 4. The blue standard curve represents the target gene and the green
standard curve represents the reference gene. A, both assays demonstrate
equivalent performance in linearity and dynamic range covering 1 µg to 1 pg.
Thus, any RNA input going forward within this range will be acceptable; B, both
assays are either saturated at the 1 µg data point or the reverse transcription
reaction is inhibited due to carryover inhibitors from the RNA sample. Consider
using less RNA (≤100 ng) or re-purifying the RNA; C, the reference assay has a
broader dynamic range than the target assay, therefore, the dynamic range is
limited. Consider reevaluating the target assay design, using less RNA (≤100 ng),
or re-purifying the RNA; D, the target assay exhibits a high standard deviation at
the lowest concentration (1 pg) and should not be considered part of the dynamic
range. This is due to a lack of sensitivity or reproducibility, and may be alleviated
by using a carrier in the RNA sample such as glycogen or non-target gDNA
carrier; E, after considering all the data, the concentration points that define the
dynamic range from rejecting the variant 1 pg data and the saturated/inhibited
1 µg data point results in an effective dynamic range (RNA loading) is 1–100 ng.
A
E
B
C
D
Cq
Initial RNA
Initial RNA
Initial RNA
Initial RNA
Initial RNA
Cq
Cq
Cq
Cq
1 pg
1 pg
1 pg
10 pg
10 pg
10 pg
100 pg
100 pg
100 pg
1 ng
1 ng
1 ng
1 µg
1 µg
1 µg
10 ng
10 ng
10 ng
10 ng
10 ng
100 ng
100 ng
100 ng
100 ng
100 ng
1 pg
1 pg
10 pg
10 pg
100 pg
100 pg
1 ng
1 ng
1 µg
1 µg
