Pcr performance not 100% efficient – Bio-Rad SsoAdvanced™ Universal Probes Supermix User Manual
Page 24
18 |
SsoAdvanced
™
Universal Probes Supermix Instruction Manual
18 |
PCR Performance Not 100% Efficient
If you have already ruled out your samples as a source for poor efficiency, then the assay may
be the cause of the problem. Please review the section on assay design in this manual for
further information (page 3).
Also, consider the following corrective action:
Perform a temperature gradient experiment to determine the optimal annealing temperature.
Set up the gradient as follows:
a. Use several representative samples in your project.
b. Set the temperature range 10ºC above and 6ºC below the calculated annealing temperature.
c. Choose the final annealing temperature based on overall performance related to specificity.
1. Evaluate the assay design by following the bioinformatics workflow outlined in the beginning
of this manual (page 3). This will help ensure that the primers are highly specific to your
target of interest and no other target region(s).
2. Perform a temperature gradient to determine the optimal annealing temperature of the
primers. Load your plate with the same reaction setup and sample for each primer set in a
column format so that you can evaluate the annealing temperatures. Set the gradient 10ºC
above and 6ºC below the calculated annealing temperature to ensure a proper temperature
range is covered. Choose the best temperature based on the overall PCR amplification,
keeping in mind that lower temperatures may reduce specificity and higher temperatures
may reduce primer binding efficiency.
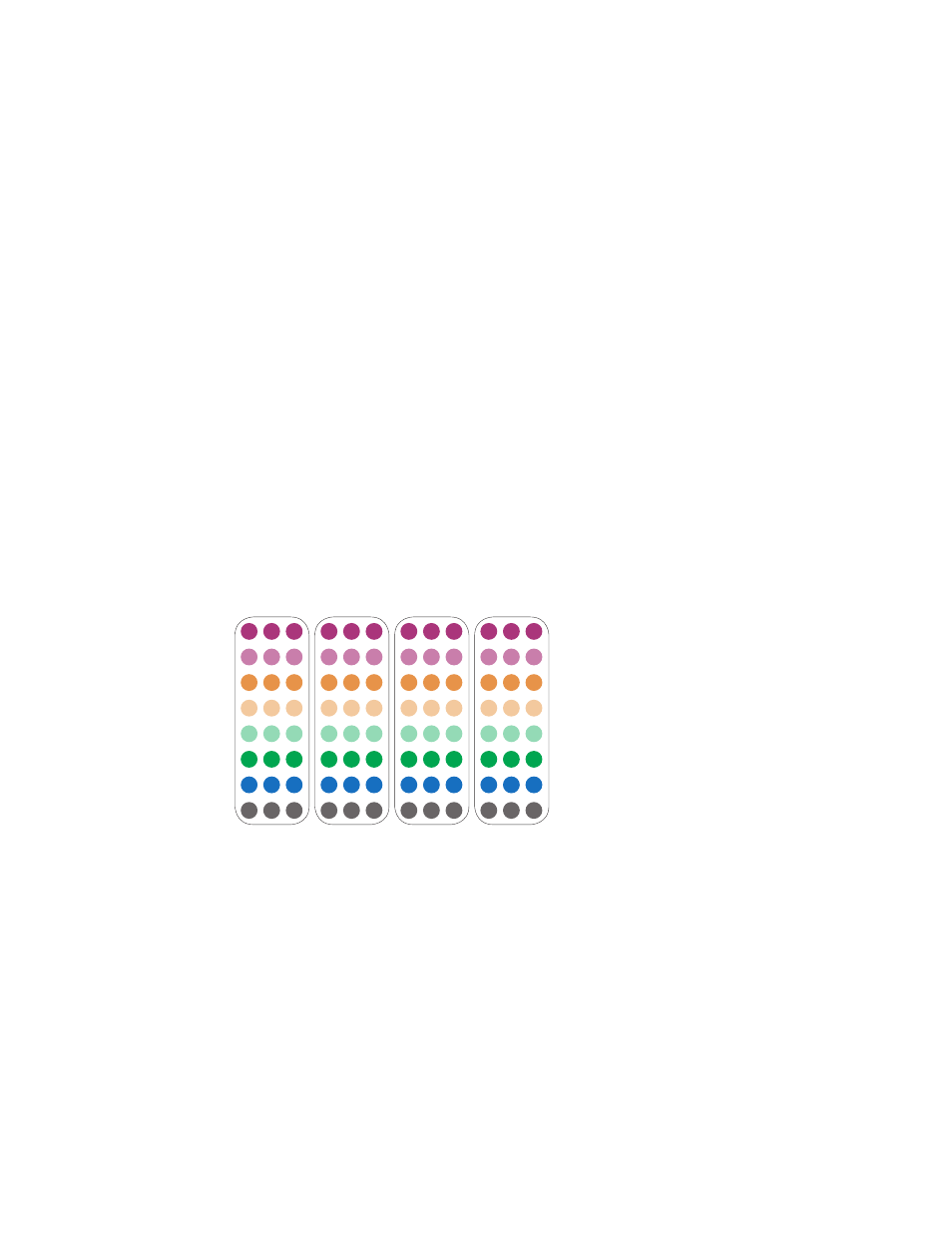
Fig. 14. Temperature gradient layout.
Primer
Set 1
Primer
Set 2
Primer
Set 3
Primer
Set 4
10°C above
Annealing T
m
= 60°C
6°C below
A 70.0
B 68.9
C 66.9
D 64.0
E 59.8
F 57.1
G 55.2
H 54.0
