Bio-Rad SsoAdvanced™ Universal Probes Supermix User Manual
Page 14
8 |
SsoAdvanced
™
Universal Probes Supermix Instruction Manual
8 |
2. From your experiment, randomly select a few samples from each group (for example,
treatments, time courses, sources) ensuring that you evaluate all variable sample groups.
3. Isolate the RNA and DNase-treat using the same protocol for all samples. Quantify and
normalize the RNA to the same concentration.
4. Perform a reverse transcription reaction for each sample using the same kit, volume, and
concentration. Dilute the cDNA, as needed, treating each sample the same to ensure there
are no differences from sample to sample in terms of volume and concentration from the
initial RNA input.
5. Perform a real-time PCR experiment using the samples and the candidate reference genes
using technical triplicates for each sample.
6. Evaluate the data for each reference gene by calculating a standard deviation for all
samples. For example, if you evaluated eight samples and seven reference genes, simply
calculate the standard deviation of those eight samples’ Cq values for each reference
gene. Thus, you will end up with seven standard deviation values. Compare the values
to determine which reference gene(s) have the lowest value. Although there is no precise
threshold for determining a good reference gene, a good rule of thumb is to ignore any
reference gene with a standard deviation higher than 0.5. If you are using a Bio-Rad CFX
real-time PCR system, you can utilize the software to automatically calculate an M-value to
assist in determining the optimal reference gene.
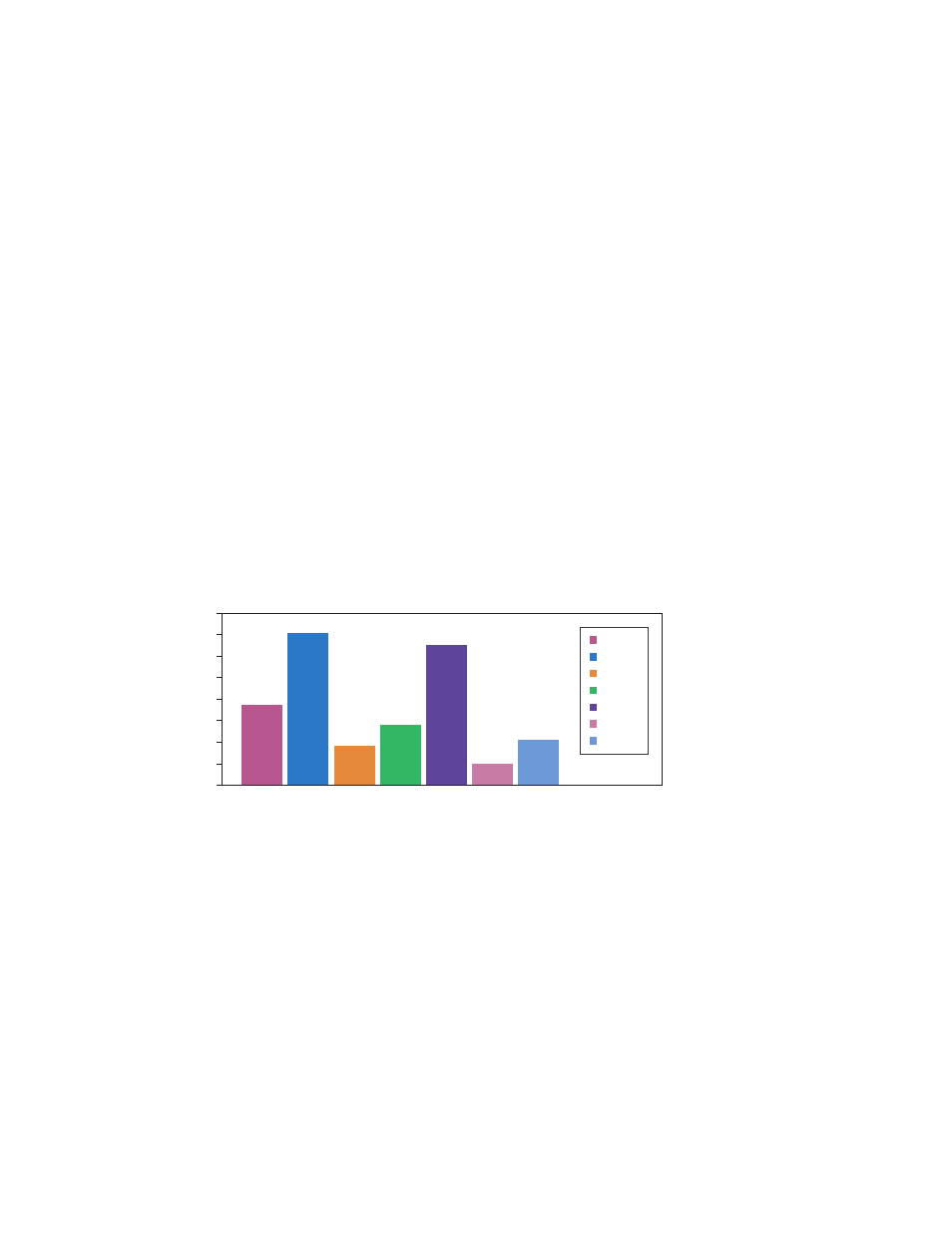
In this data set (Figure 3), TBP and PPIA are both below 0.5 and may be suitable reference
genes for the given project. Keep in mind there is no one good reference gene for all
projects, so the reference gene must be validated for every project.
Fig. 3. Seven reference genes evaluated using random samples from untreated and
treated sample groups. TBP and PPIA exhibited the lowest standard deviations with ~0.4 and
0.2, respectively. Note that GAPDH and ACTB exhibited the highest standard deviations, thus
would be unacceptable reference genes. If you are unable to find a single stable reference gene,
consider using multiple reference genes. This method involves calculating a geometric mean of
the reference gene quantities (not Cq values) prior to performing the normalization.
1.6
1.4
1.2
0.8
0.6
0.4
0.2
0
1.0
HPRT
GAPDH
TBP
18S
ACTB
PPIA
RPL13A
