Bio-Rad SsoAdvanced™ Universal Probes Supermix User Manual
Page 18
12 |
SsoAdvanced
™
Universal Probes Supermix Instruction Manual
12 |
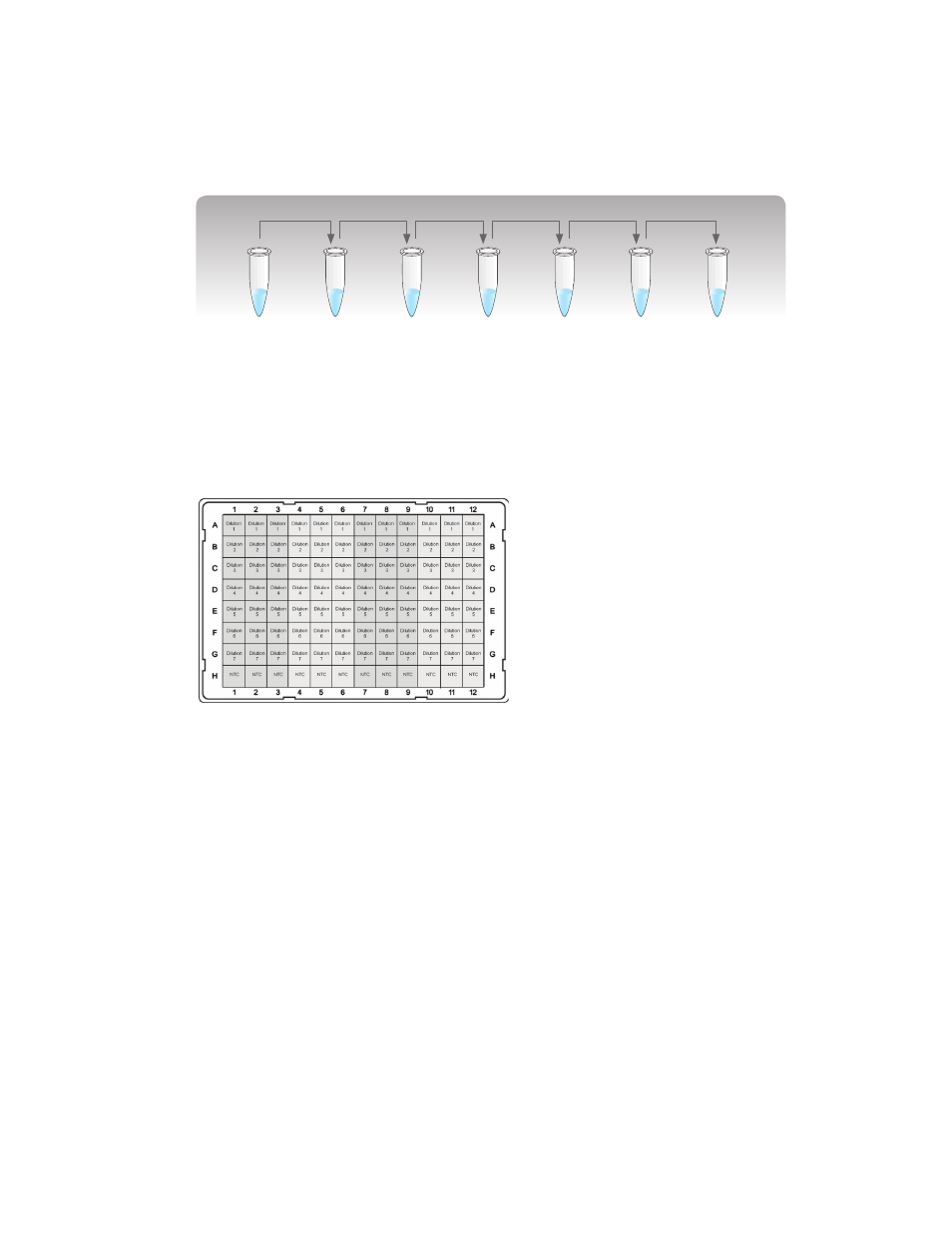
2. Prepare the real-time PCR reactions using a fresh bottle of supermix, nuclease-free water,
and primer sets. Figure 8 is an example of a plate layout.
3. Cycle according to the recommended protocol.
4. Analyze the data. Follow the guidelines in this manual for setting the baseline and threshold
prior to analyzing the data.
5. To determine which math model should be applied, simply subtract the slope value of the
reference gene from each target gene. If the ∆slope is ≤0.1, then the PCR efficiencies are
within accepted limits and the ∆∆C
T
math model can be used. If the ∆slope is ≥0.1, then the
efficiency correction math model (Pfaffl method) must be applied.
Fig. 7. Tenfold serial dilution covering 6 logs of dynamic range is prepared using a starting template
of your choice based on target expression levels.
1
1:10
1:100
1:1,000
1:10,000
1:100,000
1:1,000,000
Serial dilution of the template
1. A serial dilution of the cDNA, gDNA, or plasmid template is required to prepare the standard
curve. Ensure an adequate supply of template and an adequate volume are available to
evaluate all the assays used in the experiment.
Reference
gene
Medium
expressor
Low
expressor
High
expressor
Fig. 8. Example of a plate layout with four seven-point
standard curves with NTCs in technical triplicates —
one for each gene of interest and the reference gene.
